Team:KULeuven/Model/Cell Death
From 2008.igem.org
(Difference between revisions)
Avdmeers (Talk | contribs)
(New page: ==[https://static.igem.org/mediawiki/2008/4/47/Celldeath.zip Cell Death]== Cell Death {| border="1" |+ Parameter values Cell Death ! Name !! Value !! Refer...)
Newer edit →
(New page: ==[https://static.igem.org/mediawiki/2008/4/47/Celldeath.zip Cell Death]== Cell Death {| border="1" |+ Parameter values Cell Death ! Name !! Value !! Refer...)
Newer edit →
Revision as of 11:59, 18 July 2008
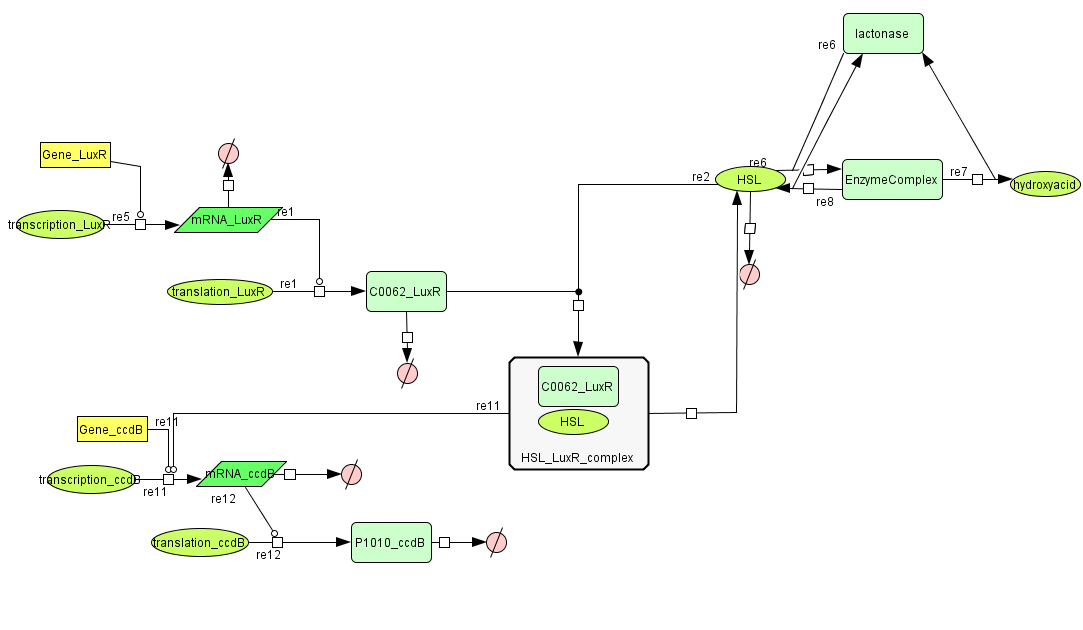
Cell Death
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| LuxR (protein) | 0.0010 /s | |
| LuxR (mRNA) | 0.00227 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| CcdB (protein) | 7.7E-5 /s | [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] |
| CcdB (mRNA) | 0.00231 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| HSL | 1.02E-6/s | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
| Dissociation Rate | ||
| ka (HSL+LuxR) | 1E6 /s | chosen to be relatively (to the other rate constants) high and such that Kd equals 1E-6 |
| kd (HSL+LuxR) | 1 /s | |
| Dissociation Constant | ||
| HSL-LuxR | 1E-6 M/L | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| Binding LuxR on LuxPromotor | 1E-9 M/L | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| Transcription Rate | ||
| LuxR (constitutive promotor) | 0.025 mRNA/s | see sections on Constitutive Promotors & E. coli transcription Rates |
| CcdB (LuxR repressor) | k_1*K_m^n/(K_m^n + [LuxR]^n) = 0.025*1E-9/(1E-9^1+[LuxR]) |
 "
"