Team:KULeuven/Model/FullModel
From 2008.igem.org
(→Full Model - Part 2) |
m (→ODE's) |
||
| Line 65: | Line 65: | ||
==== ODE's ==== | ==== ODE's ==== | ||
| + | |||
| + | todo!!! | ||
==== Parameters ==== | ==== Parameters ==== | ||
Revision as of 12:16, 2 September 2008
Contents |
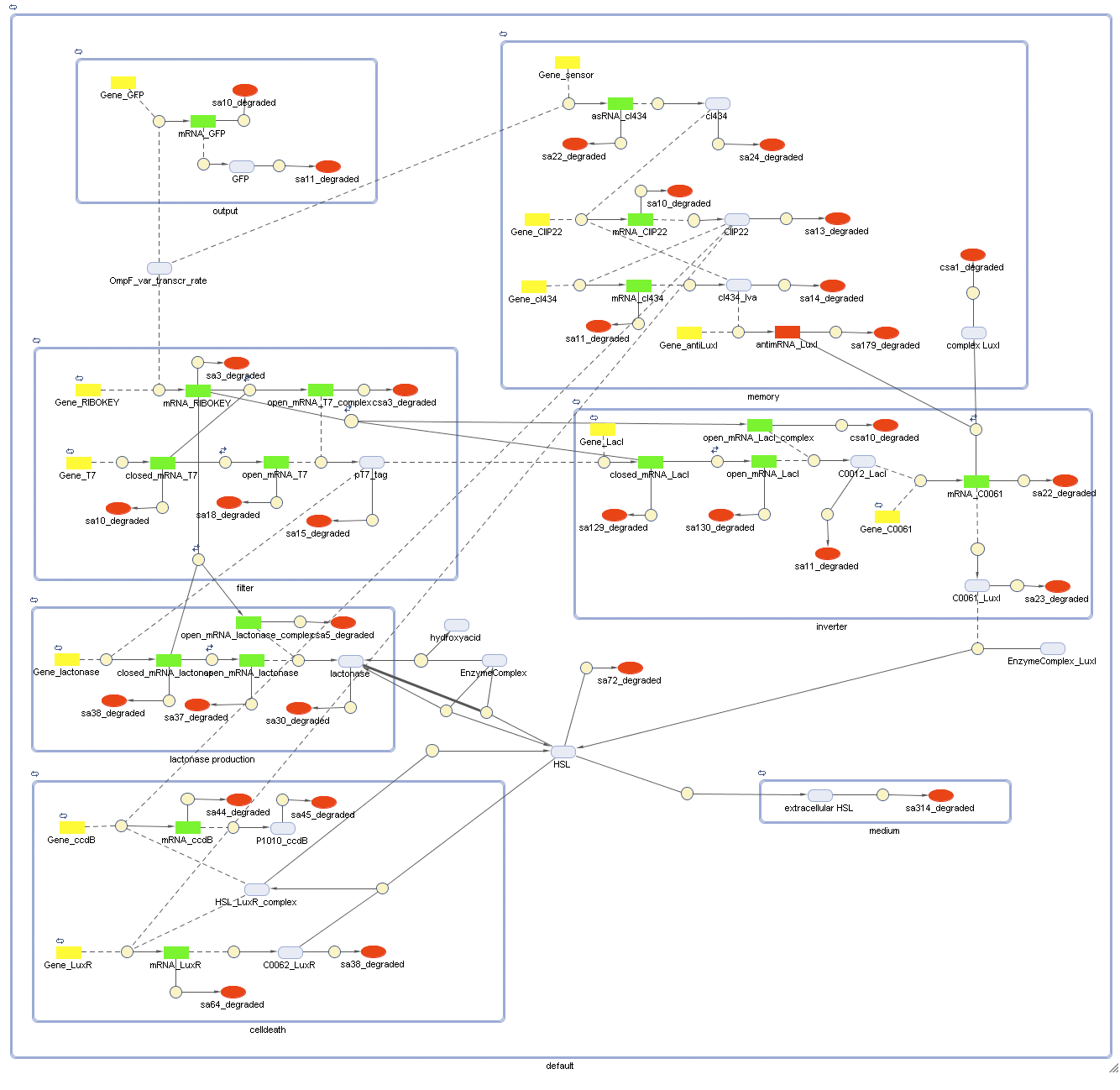
Full Model
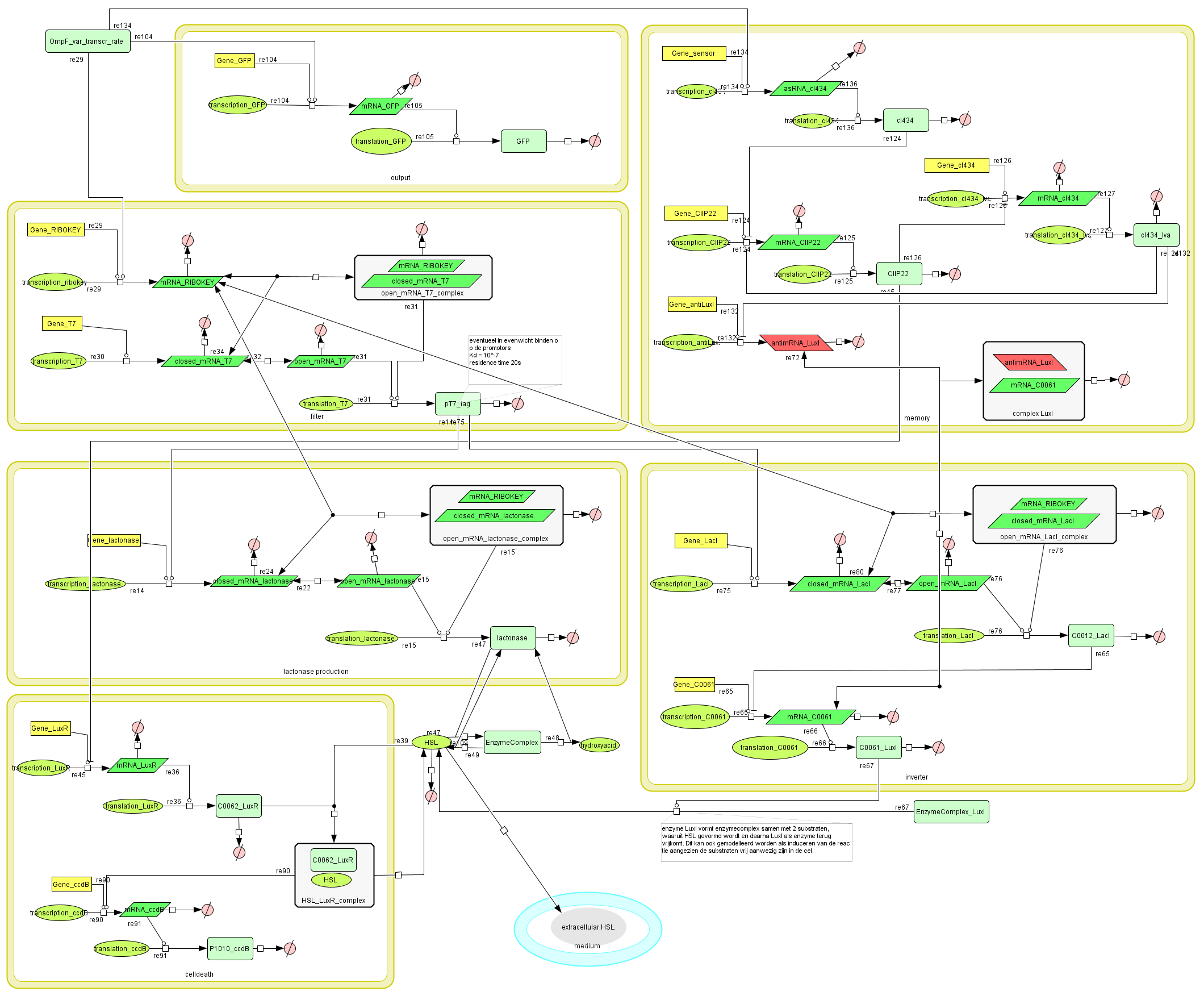
Describing the system
Dr. Coli produces a green fluorescent protein (drug) when it senses INPUT (certain disease signal in the human body). When Dr. Coli doesn't sense any INPUT anymore (the patient is cured), the invertimer initiates the molecular timer, eventually leading to cell death. Upon renewed presence of the INPUT (disease signal), the timer is reset. A filter ensures that the timer is not reset when only “noisy” INPUT signals are sensed. A memory device (stable switch) is included to ensure the clock only starts ticking when it is activated by the first input signal.
ODE's
Parameters
The parameters can be found in the sections about the simple parts of the system.
Models
CellDesigner (SBML file)
Matlab (file)
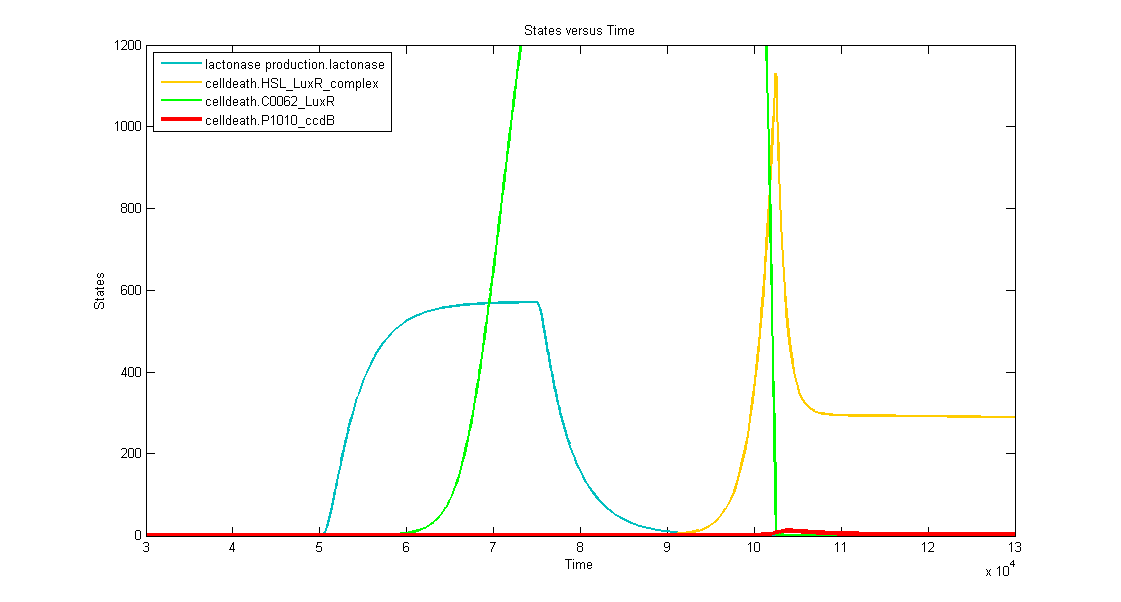
Simulations
Discussion of the simulation:
- In the beginning the memory has status "0" and the simulation clearly shows that the timer isn't activated. This means that the bacteria can grow without any problem (no ccdB-production and hence no cell death) when they haven't sensed any desease marker in the beginning.
- At t=50.000s the system is activated by a long lightpulse (simulating the insertion of the bacteria in the patient's body and sensing desease marker for the first time). This results in:
- switching of the memory to status "1" --> activation of transcription of LuxR (green curve)
- activation of the filter (desease marker is strong enough) which is reflected by the production of lactonase (blue curve), situated just after the filter mechanism.
- At t=75.000 the lightpulse is switched of (simulating that there's no more desease marker: the patient is cured). This results in:
- stopping the production of lactonase, which starts to degrade naturally (blue curve)
- stopping the production of LacI, which stops repressing the transcription of LuxI. In this way HSL can be produced, but, since there's still lactonase present in the cell, there won't be a visible increase in HSL-LuxR-complex because the HSL is being transformed into hydroxyacid.
- At t~=92.000 the lactonase is almost totally degraded which enables the HSL to form a complex with LuxR (yellow curve). This means that we have a timer that starts with a delay of +- 17.000s. At this point the HSL-LuxR-complex really starts to build up gradually (TIMER) for about 10.000s.
- At t~=102.000s the HSL-LuxR-complex peaks, with at the same time (free) LuxR decreasing to almost zero.
- At this point all the newly produced LuxR will go into complex with HSL and no free LuxR will remain. From this point on, the HSL-LuxR-complex will decrease to a steady state value which is equal to the production rate of LuxR-proteins.
- BUT you can see clearly that the ccdB (red curve) also peaks (to a value of 10 to 20 molecules) which is enough to result in cell death.
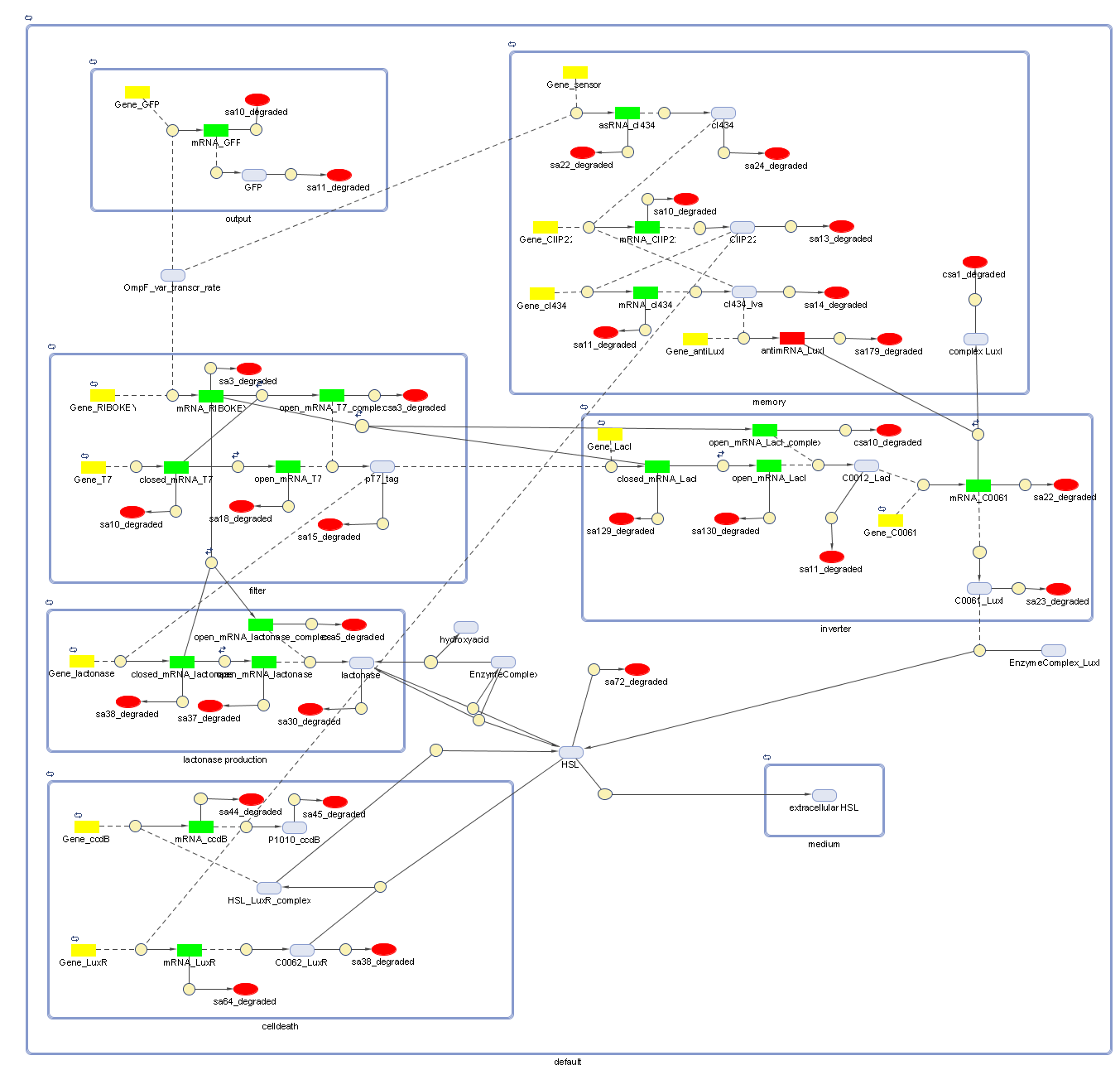
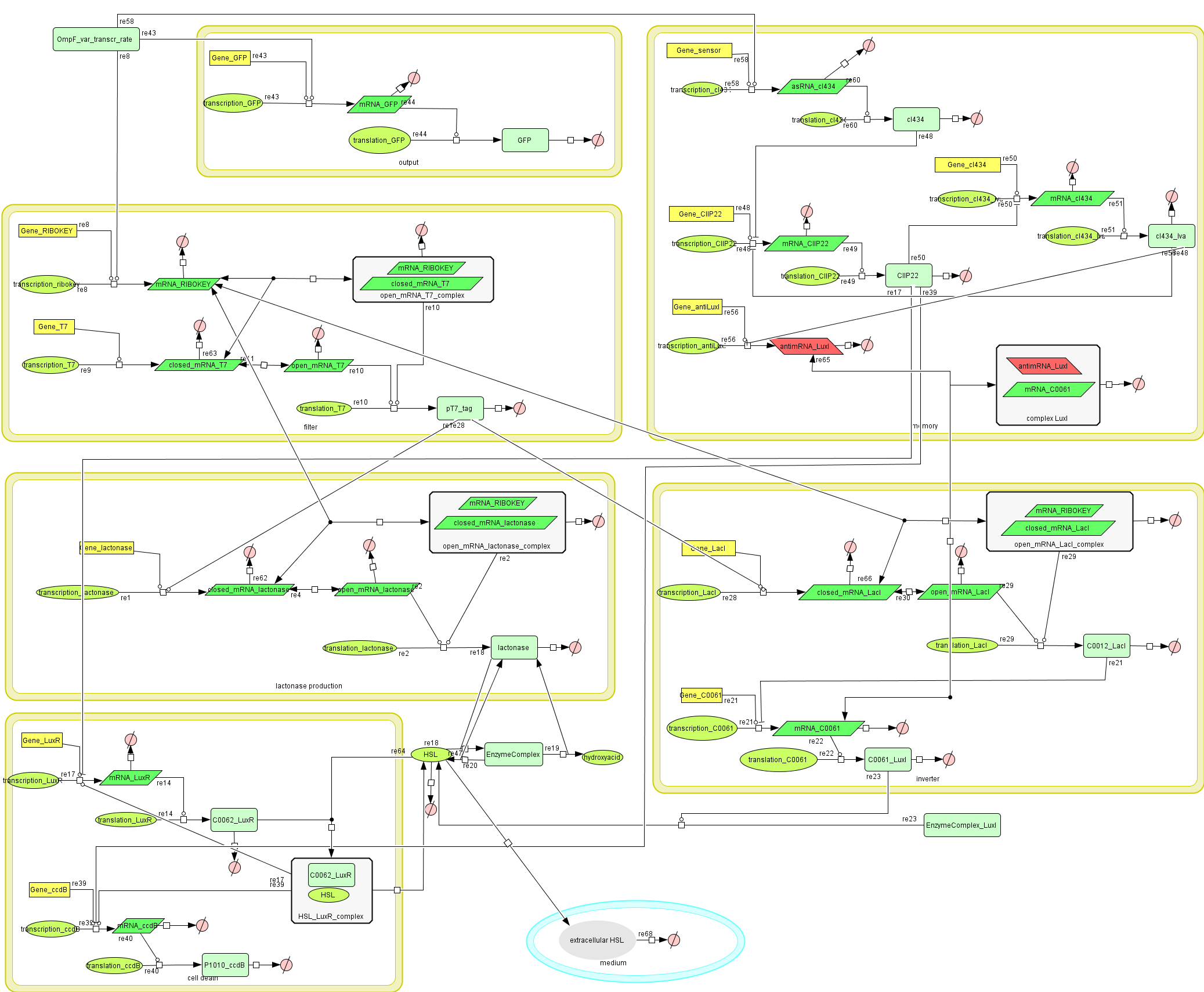
Full Model - Part 2
Extension to the previous model
During the summer we switched from the above system to a new one which you can see just below. These changes have been documented in Model:New Cell Death.
Describing the system
ODE's
todo!!!
Parameters
The parameters can be found in the sections about the simple parts of the system.
Extension: parameters of HSL-LuxR auto-activation can be found in Model:New Cell Death.
 "
"