Team:KULeuven/Software/MultiCell
From 2008.igem.org
(→Step 1: Load Project) |
m |
||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Scripting}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| Line 52: | Line 54: | ||
2. The cell division process is very simplified in this toolbox. It keeps track of the age of the cells and when the time is right for division, the mothercell is divided in 2 new cells: a 'reïncarnation' of the mothercell (with adjusted initial conditions) and a new cell (with initial conditions adjusted to the mothercell). In this stage of the toolbox we don't take into account the possibility of cell growth or other events during the cell division process (extensions in a future release...). | 2. The cell division process is very simplified in this toolbox. It keeps track of the age of the cells and when the time is right for division, the mothercell is divided in 2 new cells: a 'reïncarnation' of the mothercell (with adjusted initial conditions) and a new cell (with initial conditions adjusted to the mothercell). In this stage of the toolbox we don't take into account the possibility of cell growth or other events during the cell division process (extensions in a future release...). | ||
| - | + | <html><center><div style="float:center;background:#5cc7dd;width:600px;height:170px" > | |
| + | <p> | ||
| + | The MultiCell Toolbox is made available for non commercial research purposes only under the GNU General Public License. However, notwithstanding any provision of the <a href="http://www.gnu.org/copyleft/gpl.html"> GNU General Public License</a>, MultiCell Toolbox may not be used for commercial purposes without explicit written permission after contacting <a href="mailto:igem@kuleuven.be?SUBJECT=IGEM 2008:License MultiCell Toolbox">igem@kuleuven.be</a>. | ||
| + | </p> | ||
| + | <p> When reporting results obtained by MultiCell Toolbox, one should refer to </p> | ||
| + | <p> <center>iGEM2008/Team:KULeuven, Nick Van Damme <br> | ||
| + | MultiCell Toolbox </center> | ||
| + | </p> | ||
| + | </div></center></html> | ||
== File == | == File == | ||
| - | + | [https://static.igem.org/mediawiki/2008/f/ff/MultiCell_Toolbox.zip Zip-File] | |
| + | |||
| + | To start the GUI: open GUI_start.fig or type GUI_start in MATLAB | ||
Latest revision as of 15:03, 3 October 2008
Contents |
Introduction
Have you ever wanted to simulate the effects of your MATLAB Symbiology project in a multicellular environment? This toolbox enables you to expand your 'what happens in 1 cell'-model into a network of interconnected cells with the full abilities of a growing population of cells. It delivers you an easy-to-use graphical interface to configure the simulation settings and with just 1 click an extensive simulation is started.
The Program
GUI
Use
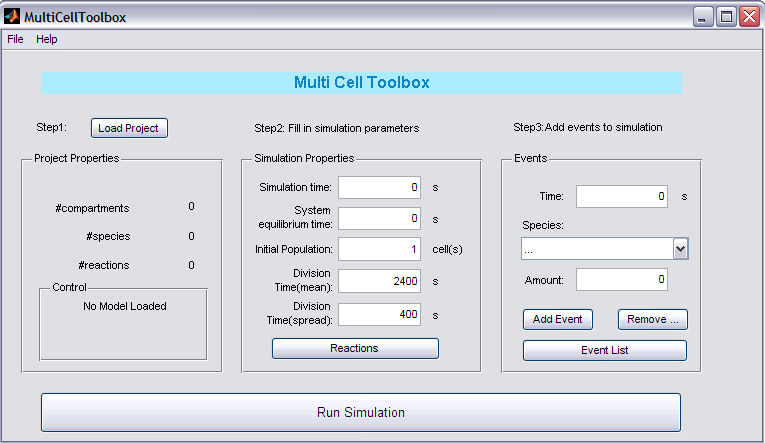
Step 1: Load Project
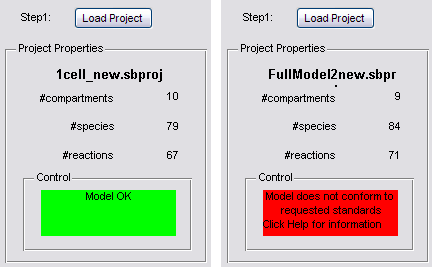
Push the button to load your '1cell'-project. In the Project Properties panel the name of your project will appear together with some crucial data about your project: the number of compartments, species and reactions. After loading your project, it will be checked to conform to the requested standards of the toolbox (see Remarks) and a note will appear in the Control panel.
Step 2: Fill in simulation parameters
In the Simulation Properties panel you can enter all the necessary parameters for the simulation:
- Simulation Time: duration of the simulation (with cell division action)
- System Equilibrium time: time the system needs to reach its equilibrium state (first simulation starts with Species.InitialCondition equal to their initial condition in the project)
- Initial Population: the amount of cells before the simulation starts (with cell division action)
- Division Time (mean): mean time for 1 cell till (next) division
- Division Time (spread): possible spread around the mean time for 1 cell till (next) division
You also need to enter the reactions that involve species in 'cell1' and have to be copied to their daughter cells. This can be done by clicking the Reactions button and selecting these reactions.
Step 3: Add events to simuation
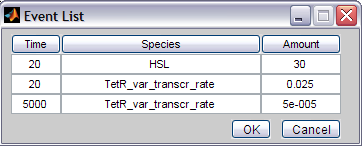
One of the pros of this toolbox is the included possibility of introducing events (just like in MATLAB). Select the starting time (trigger), the specie involved and the amount of the specie and then click 'Add...'. The other buttons also speak for themselves.
Step 4: RUN
After configuring all the necessary settings, you're now ready to run the application!
Remarks
1. The loaded projectfile needs to conform to some standards to be able to simulate with the Multi Cell Toolbox:
- There must be a compartment with the name 'system', which encomprises everything.
- There must be a compartment with the name 'cell1', which represents the basic cell that will be used for cell replication/division. Its 'Owner' must be the 'system'-compartment.
2. The cell division process is very simplified in this toolbox. It keeps track of the age of the cells and when the time is right for division, the mothercell is divided in 2 new cells: a 'reïncarnation' of the mothercell (with adjusted initial conditions) and a new cell (with initial conditions adjusted to the mothercell). In this stage of the toolbox we don't take into account the possibility of cell growth or other events during the cell division process (extensions in a future release...).
The MultiCell Toolbox is made available for non commercial research purposes only under the GNU General Public License. However, notwithstanding any provision of the GNU General Public License, MultiCell Toolbox may not be used for commercial purposes without explicit written permission after contacting igem@kuleuven.be.
When reporting results obtained by MultiCell Toolbox, one should refer to
MultiCell Toolbox
File
To start the GUI: open GUI_start.fig or type GUI_start in MATLAB
 "
"