Team:KULeuven/Software/MultiCell
From 2008.igem.org
| Line 1: | Line 1: | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | == | + | == Introduction == |
| + | |||
| + | Have you ever wanted to simulate the effects of your MATLAB Symbiology project in a multicellular environment? This toolbox enables you to expand your 'what happens in 1 cell'-model into a network of interconnected cells with the full abilities of a growing population of cells. It delivers you an easy-to-use graphical interface to configure the simulation settings and with just 1 click an extensive simulation is started. | ||
| + | |||
| + | == The Program == | ||
| + | |||
| + | === GUI === | ||
[[Image:MultiCellToolbox.PNG|center]] | [[Image:MultiCellToolbox.PNG|center]] | ||
| + | |||
| + | === Use === | ||
| + | |||
| + | ==== Step 1: Load Project ==== | ||
| + | |||
| + | Push the button to load your '1cell'-project. In the Project Properties panel the name of your project will appear together with some crucial data of your project: the number of compartments, species and reactions. After loading your project, it will be checked for conforming to the requested standards of the toolbox (see [https://2008.igem.org/Team:KULeuven/Software/MultiCell#Remarks Remarks]) and a note will appear in the Control panel. | ||
| + | |||
| + | ==== Step 2: Fill in simulation parameters ==== | ||
| + | |||
| + | In the Simulation Properties panel you can enter all the necessary parameters for the simulation: | ||
| + | * Simulation Time: duration of the simulation (with cell division action) | ||
| + | * System Equilibrium time: time the system needs to reach its equilibrium state (first simulation starts with Species.InitialCondition equal to their initial condition in the project) | ||
| + | *Initial Population: the amount of cells before the simulation starts (with cell division action) | ||
| + | *Division Time (mean): mean time for 1 cell till (next) division | ||
| + | *Division Time (spread): possible spread around the mean time for 1 cell till (next) division | ||
| + | |||
| + | You also need to enter the reactions that involve species in 'cell1' and have to be copied to their daughter cells. This can be done by clicking the Reactions button and selecting these reactions. | ||
| + | |||
| + | ==== Step 3: Add events to simuation ==== | ||
| + | |||
| + | One of the pros of this toolbox is the included possibility of introducing events (just like in MATLAB). Select the starting time (trigger), the specie involved and the amount of the specie and then click 'Add...'. The other buttons also speak for themselves. | ||
| + | |||
| + | ==== Step 4: RUN ==== | ||
| + | |||
| + | After configuring all the necessary settings, you're now ready to run the application! | ||
| + | |||
| + | |||
| + | == Remarks == | ||
| + | |||
| + | |||
| + | == File == | ||
| + | |||
| + | COMING SOON | ||
| + | |||
| + | == Features == | ||
Revision as of 09:53, 9 September 2008
Contents |
Introduction
Have you ever wanted to simulate the effects of your MATLAB Symbiology project in a multicellular environment? This toolbox enables you to expand your 'what happens in 1 cell'-model into a network of interconnected cells with the full abilities of a growing population of cells. It delivers you an easy-to-use graphical interface to configure the simulation settings and with just 1 click an extensive simulation is started.
The Program
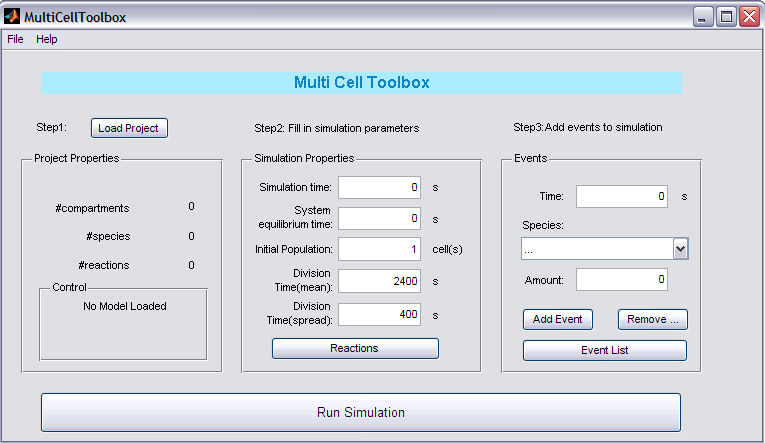
GUI
Use
Step 1: Load Project
Push the button to load your '1cell'-project. In the Project Properties panel the name of your project will appear together with some crucial data of your project: the number of compartments, species and reactions. After loading your project, it will be checked for conforming to the requested standards of the toolbox (see Remarks) and a note will appear in the Control panel.
Step 2: Fill in simulation parameters
In the Simulation Properties panel you can enter all the necessary parameters for the simulation:
- Simulation Time: duration of the simulation (with cell division action)
- System Equilibrium time: time the system needs to reach its equilibrium state (first simulation starts with Species.InitialCondition equal to their initial condition in the project)
- Initial Population: the amount of cells before the simulation starts (with cell division action)
- Division Time (mean): mean time for 1 cell till (next) division
- Division Time (spread): possible spread around the mean time for 1 cell till (next) division
You also need to enter the reactions that involve species in 'cell1' and have to be copied to their daughter cells. This can be done by clicking the Reactions button and selecting these reactions.
Step 3: Add events to simuation
One of the pros of this toolbox is the included possibility of introducing events (just like in MATLAB). Select the starting time (trigger), the specie involved and the amount of the specie and then click 'Add...'. The other buttons also speak for themselves.
Step 4: RUN
After configuring all the necessary settings, you're now ready to run the application!
Remarks
File
COMING SOON
 "
"