Team:Lethbridge CCS/Notebook/October
From 2008.igem.org
(→18 Oct 2008) |
(→16 Oct 2008) |
||
| Line 130: | Line 130: | ||
[[Image:pSB1A2_LIC_pur_gel.jpg|pSB1A2_LIC_pur_gel|250px]] [Lanes: 1/25 dilution of 1/10 #1, 1kb ladder (punctured well?), 1/10 #1, 1/10 #2, 1/100 #1, 1/100 #2, 1kb ladder, 1/25 dilution of 1/10 #2] | [[Image:pSB1A2_LIC_pur_gel.jpg|pSB1A2_LIC_pur_gel|250px]] [Lanes: 1/25 dilution of 1/10 #1, 1kb ladder (punctured well?), 1/10 #1, 1/10 #2, 1/100 #1, 1/100 #2, 1kb ladder, 1/25 dilution of 1/10 #2] | ||
| - | * All plasmid preps seem to have worked; circular plasmid is running a little below 2000bp, which seems a little small but is probably OK. | + | * All plasmid preps seem to have worked; circular plasmid is running a little below 2000bp, which seems a little small but is probably OK. May be due to insufficiently resolved gel. |
=== 17 Oct 2008 === | === 17 Oct 2008 === | ||
Revision as of 01:15, 19 October 2008
For earlier work, see September Notebook, or go back to Main CCS Notebook.
Contents |
4 Oct 2008
(Marc, Glenda, Nathan)
Met to discuss why we had not obtained green colonies from our transformation as expected. There was definitely cutting happening from the EcoRV digest, but the bands showed up at the wrong size. Final conclusion was that probably nothing had worked right from step 1! Most likely what we thought was pSB1A7 actually wasn't, a conclusion we came to based on our own funny results, and on some of the results obtained by the U of L team. So we've probably just been seeing the original template plasmid in all our follow up gels, etc.
Decided to start again, this time using pSB1A2 (though it doesn't have the double terminators we would actually like to see), as the U of L team has it and knows it works. So new primers were designed for this, as well as for xylE, a gene the ULeth team is using, which we hope to use to see whether we can really build a 'new' BioBrick using our method.
6 Oct 2008
(Peter, Elizabeth, Glenda, Nathan)
Reviewed Saturday's discussion; checked that primers were correct and sent off order; discussed our logo and ordering of T-shirts; reviewed steps we would have to go through once our new primers arrive
11 Oct 2008
(Peter, Elizabeth, Marc, Glenda)
- Roxanne had already resuspended and diluted our primers (psB1A2 forward and reverse; LIC_xylE forward and reverse) [100 μM stock; 10μM working].
- Set up and ran PCRs on pSB1A2 and LIC_xylE
- for each, we did 1 x, 1/10 x and 1/100 x of our template and a range of annealing temperatures (54°C, 58°C, 62°C for pSB1A2; 48°C, 52°C, 56°C for LIC_xylE), giving nine variations for each PCR;
- used materials and amounts as per Phusion polymerase kit
- Cycling temperatures and times for PCR
Initial denaturation 98°C 30 sec ___
Denaturation 98°C 10 sec |
Annealing gradient as above 30 sec |--- 30 cycles
Extension 72°C 1 min ___|
Final extension 72°C 10 min
Hold 4°C hold
- numbering:
54°C 58°C 62°C
1x 1 4 7
1/10x 2 5 8
1/100x 3 6 9
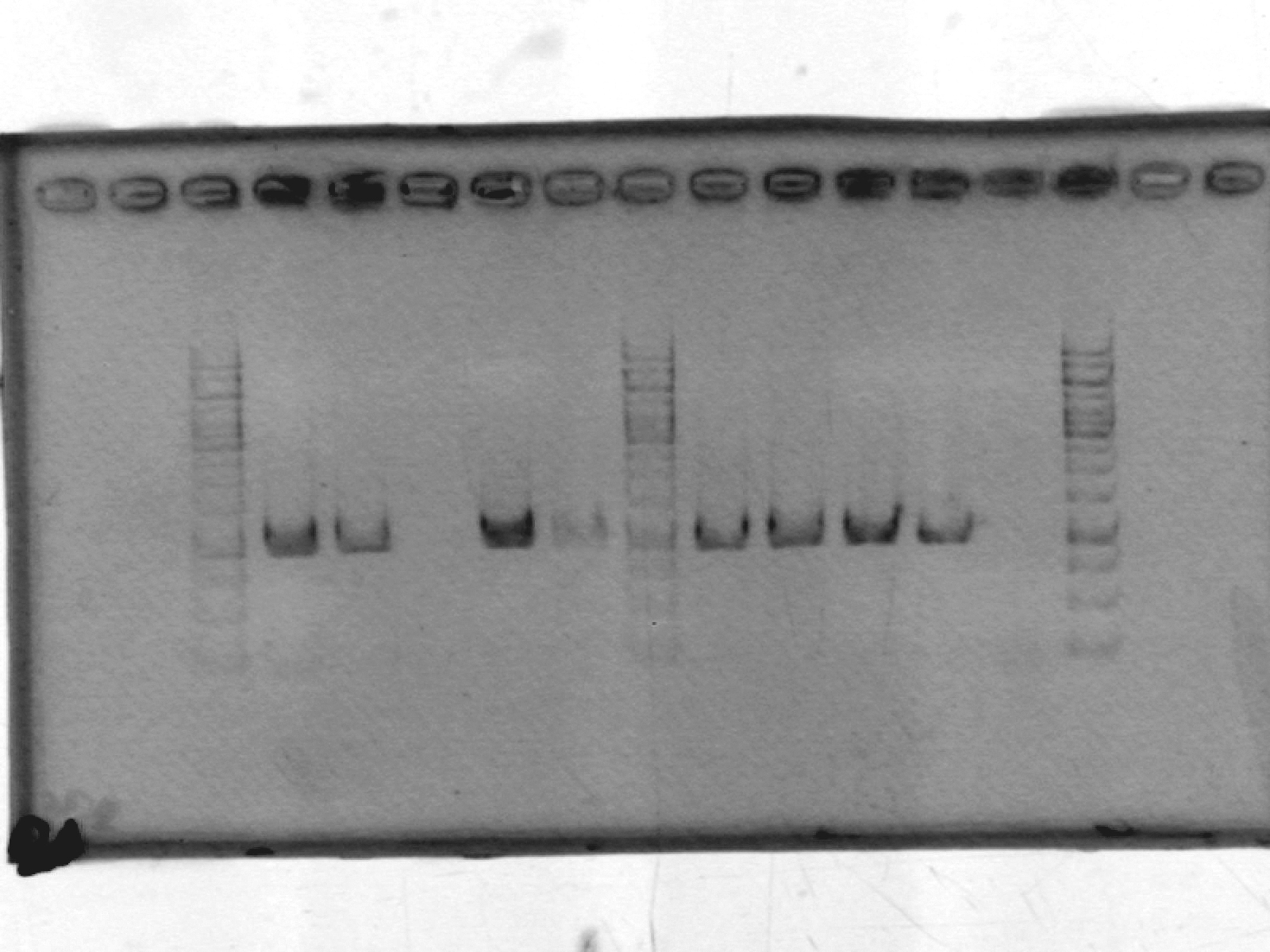
- ran a 1% agarose gel on pSB1A2_LIC; expected size ~2000 bp. No results. After discussion, we think this may be due to erroneously putting in only enough polymerase for one reaction, not 10.
 [lanes: 1kb ladder, 1-5, 1kb ladder, 6-9, blank, 1kb ladder]
[lanes: 1kb ladder, 1-5, 1kb ladder, 6-9, blank, 1kb ladder]
- ran a 1% agarose gel on xylE_LIC; expected size ~1000bp.
 [lanes: 1kb ladder, 1-5, 1kb ladder, 6-9, blank, 1kb ladder]
[lanes: 1kb ladder, 1-5, 1kb ladder, 6-9, blank, 1kb ladder]
13 Oct 2008
(Peter, Marc - yes, we were in the lab on Thanksgiving!)
- Re-did PCR of pSB1A2, this time using GFP complete (1) plasmid-prepped by Roxanne.
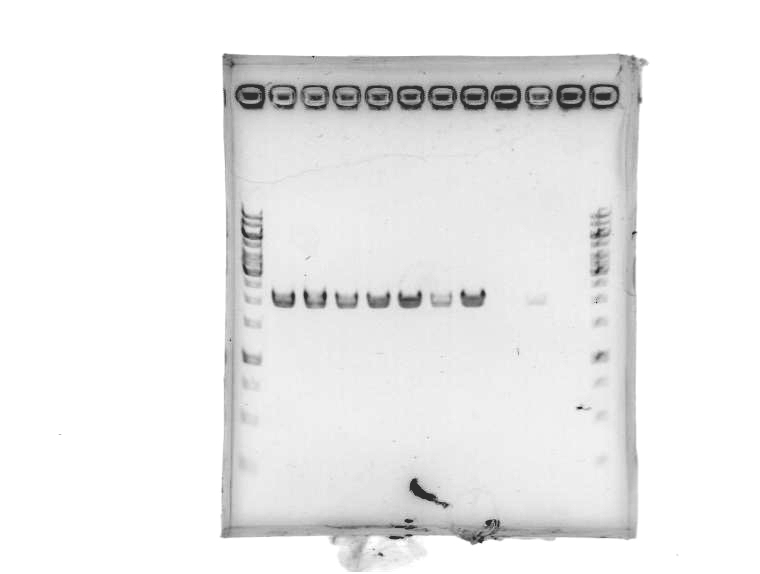 [Lanes: 1kb ladder, 1-9, blank, 1kb ladder]
[Lanes: 1kb ladder, 1-9, blank, 1kb ladder]
- Seems to have worked. Possible problem: circular pSB1A2 with GFP complete (~3000bp) may run at the same place as linear pSB1A2 alone (~2000bp). Will run another gel tomorrow to check.
14 Oct 2008
(Marc)
- Ran 1% agarose gel to check where pSB1A2 with GFP complete (template for our pSB1A2 PCR) runs on a gel.
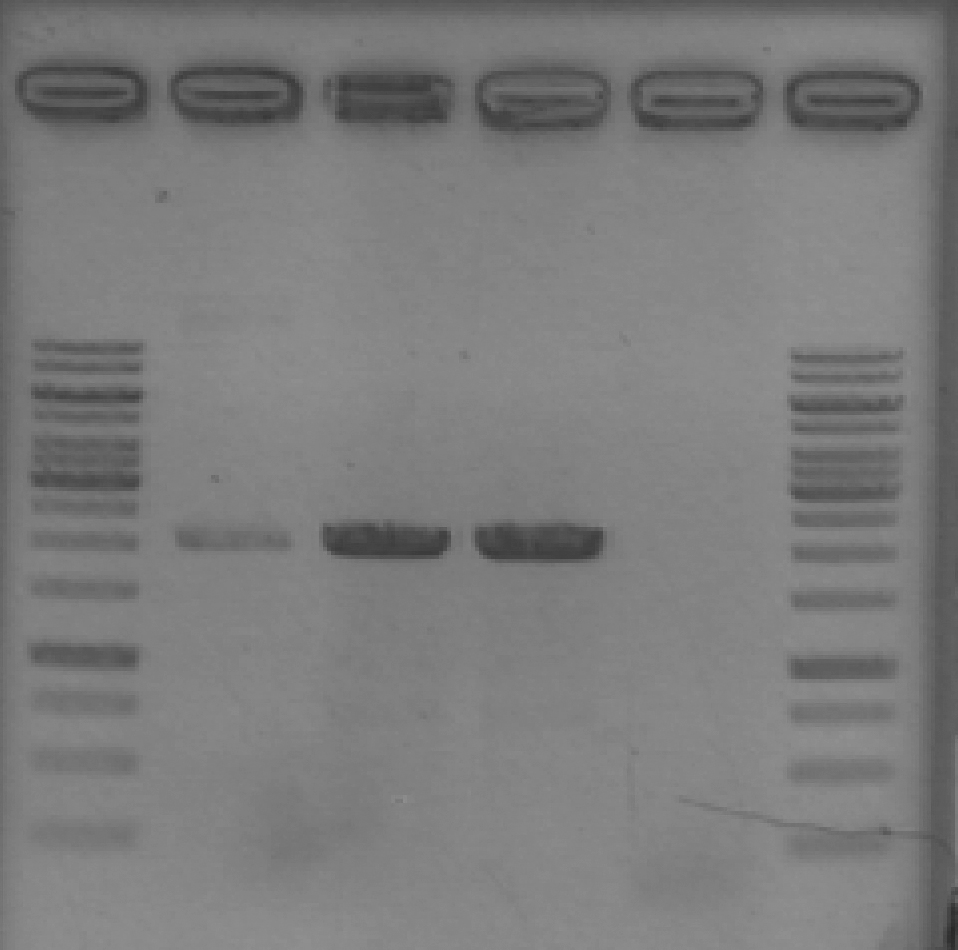 [Lanes: 1kb ladder, GFP complete (1), #2, #5, #8, 1kb ladder]
[Lanes: 1kb ladder, GFP complete (1), #2, #5, #8, 1kb ladder]
- Unfortunately, template (circular) runs at the same place as our PCR product. We could possibly digest the BioBrick plasmid first; then at least there wouldn't be any competition for transformation.
(Elizabeth, Peter, Glenda)
- Used Fermentas Rapid Ligation kit on our around the world PCR products(1/10, 1/100 concentration, 54°)
-1 μL PCR product 2 μL 5x buffer incubated for 1 hour at room temperature 1 μL ligase 6 μL Water
- Did a transformation of ligated DNA into highly competent cells (using 1 μL ligation mix (1/10, 1/100, pUC19 control)
- left on ice for 30 min
- heat-shock for 30 sec at 42 degrees
- ice for 2 minutes
- add 900 μL SOC medium, 1 hour in shaker at 37 degrees
- plated concentrated and dilute of each of above dilutions
- incubated overnight
- At the U of L iGEM meeting, we discussed our concern about template running at same place as PCR product. HJ recommended doing a DpnI digest since this would cleave the template but leave our PCR product intact. We could then run a gel to compare sizes.
15 Oct 2008
(Glenda, Elizabeth, Peter)
- Interestingly enough, where we had expected to see all or almost all green colonies (if template was present as expected), but in fact there were only white colonies
- Grew up colonies (2 each from our 1/10 and 1/100 dilute plates)
- 5 μL ampicillin, add with colonies to culture
- Incubate at 37°C for 12 hours
- Set up DpnI digestion
15 μL PCR product/template (used 1x LIC_GFP) 1.75 μL 10x Tango buffer 1 μL DpnI
- Incubated 1 hour at 37°C and then ran a gel
Lane 1: 1 kb GeneRuler ladder Lane 2: GFP with DpnI Lane 3: GFP without DpnI Lane 4: LIC_GFP with DpnI Lane 5: LIC_GFP without DpnI Lane 6: 1 kb GeneRuler ladder
- gel looked a little strange (not well-defined), but seems to verify that our early amplification worked; Lane 2 had marks at a variety of lengths, indicating cutting of GFP with DPN, while the other lanes all looked very similar,with only one length, indicating there was very little template left in our LIC_GFP. The only slightly puzzling thing was the length of the DNA it seemed to show (shorter than expected)
16 Oct 2008
(Elizabeth, Marc)
- Performed miniprep of pSB1A2_LIC from 'LIC_GFP' cultures, as per QIAprep Spin Kit.
- Notes: did not do optional PB buffer step; pellet of 1/100 #2 was very thin and elongated.
- Prepared glycerol stocks of all four cultures. Stored in HJ's -80°C
- Quantified resulting plasmid concentrations using UV/vis spectrophotometer at 1:25 dilution.
Sample Concentration (ng/μL) 260/280nm absorbance ratio 1/10 #1 57 1.83 1/10 #2 81 1.83 1/100 #1 88 1.95 1/100 #2 50 1.90
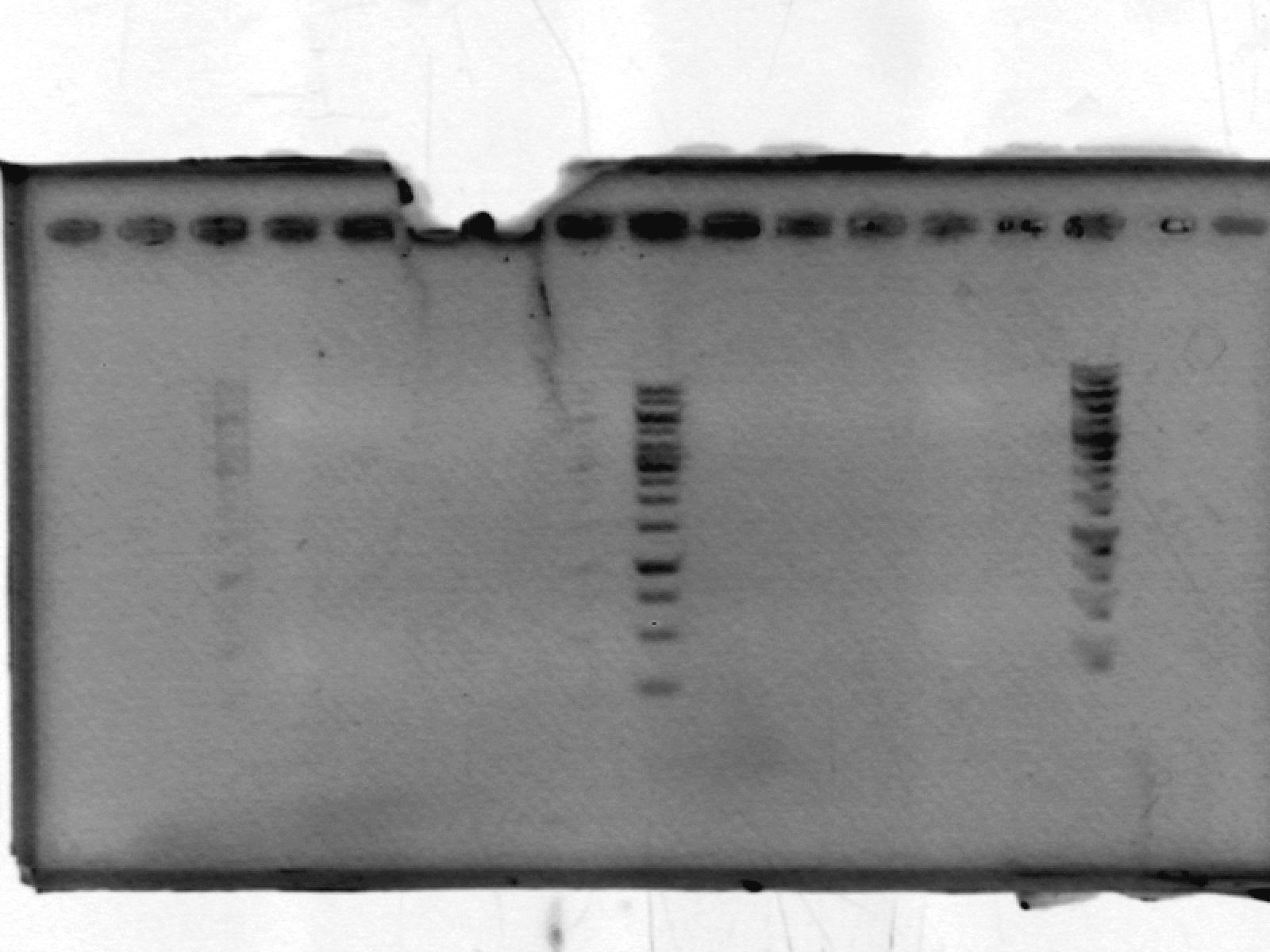
- Ran 1% agarose gel to check size of purified plasmid (100V, 25min)
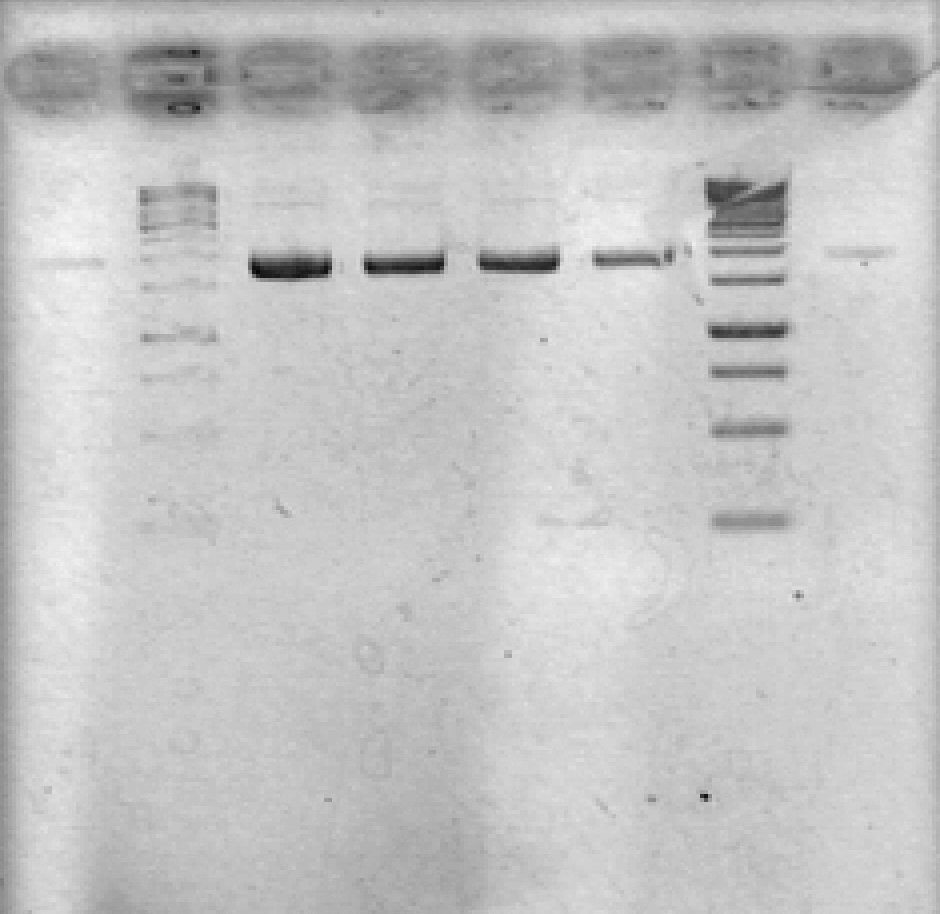 [Lanes: 1/25 dilution of 1/10 #1, 1kb ladder (punctured well?), 1/10 #1, 1/10 #2, 1/100 #1, 1/100 #2, 1kb ladder, 1/25 dilution of 1/10 #2]
[Lanes: 1/25 dilution of 1/10 #1, 1kb ladder (punctured well?), 1/10 #1, 1/10 #2, 1/100 #1, 1/100 #2, 1kb ladder, 1/25 dilution of 1/10 #2]
- All plasmid preps seem to have worked; circular plasmid is running a little below 2000bp, which seems a little small but is probably OK. May be due to insufficiently resolved gel.
17 Oct 2008
(Glenda, Elizabeth, Peter)
- Set up EcoRV digest of purified pSB1A2_LIC plasmid (Fermentas)
- components and amounts (for each of 4 colonies grown up previously)
DNA 5 μL
Buffer 1 μL incubated overnight
EcoRV 1 μL
Water 3 μL
18 Oct 2008
(Paul, Peter, Elizabeth, Marc)
- Ran 1% agarose gel to check results of yesterday's EcoRV digest. (2 μL loading volumes; 5 μL ladder; 100V for 35 min)
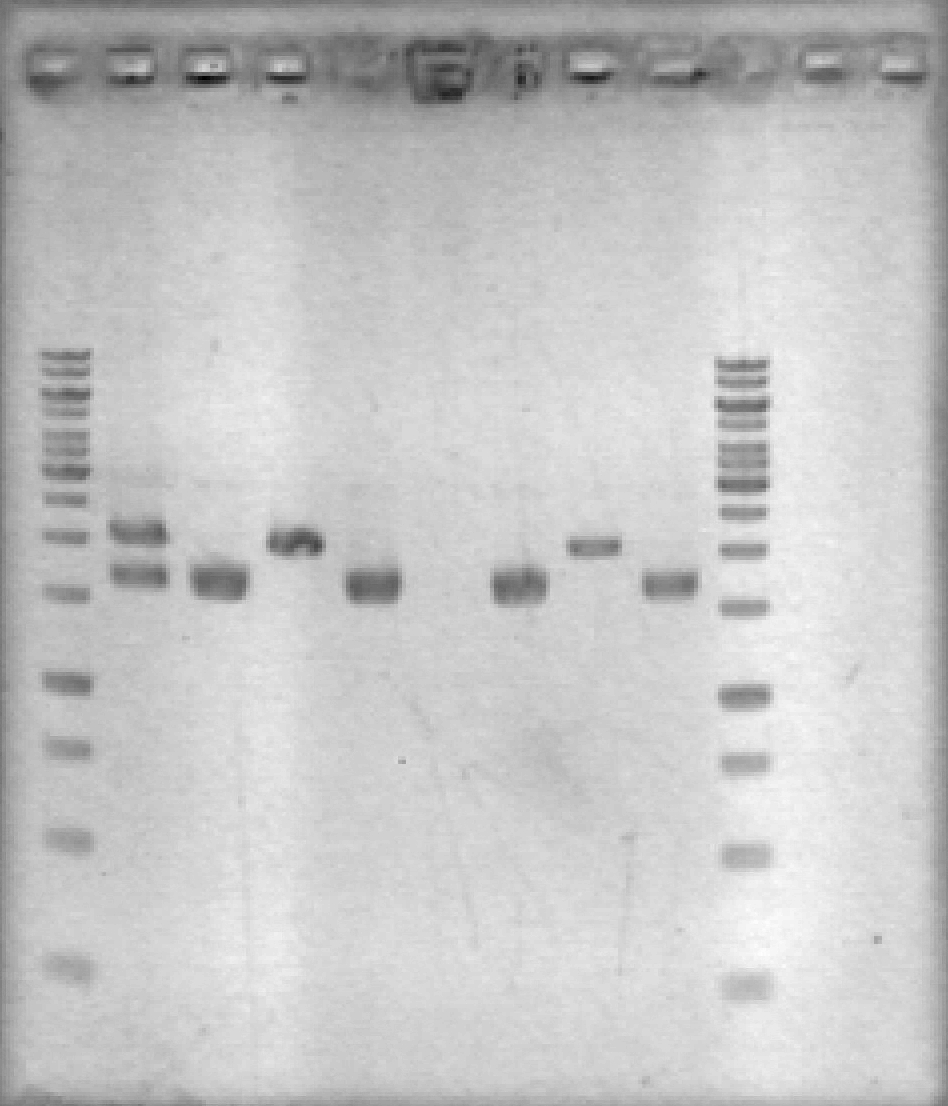 [Lanes: 1kb ladder, 1/10 #1 Cut & Uncut, 1/10 #2 Cut & Uncut, 1/100 #1 Cut & Uncut, 1/100 #2 Cut & Uncut, 1kb ladder]
[Lanes: 1kb ladder, 1/10 #1 Cut & Uncut, 1/10 #2 Cut & Uncut, 1/100 #1 Cut & Uncut, 1/100 #2 Cut & Uncut, 1kb ladder]
- Some strange results, but there was definitely digestion. It seems that 1/10 #1 may not have been fully digested; no idea why the uncut 1/100 #1 didn't show up. The #2 samples, which look 'normal', will be used for subsequent steps.
- NB: uncut plasmid is around 1700bp, which is a little smaller than it looked on the previous gel. Cut (linear) plasmid is right around 2000bp where it belongs.
- Performed T4 digestion on EcoRV-cut pSB1A2_LIC, and on PCR-amplified xylE_LIC and GFP_LIC. Had enough
Main Page | Team | Project | Lab Notebook
 "
"