Team:Michigan/Project
From 2008.igem.org
|
|---|
|
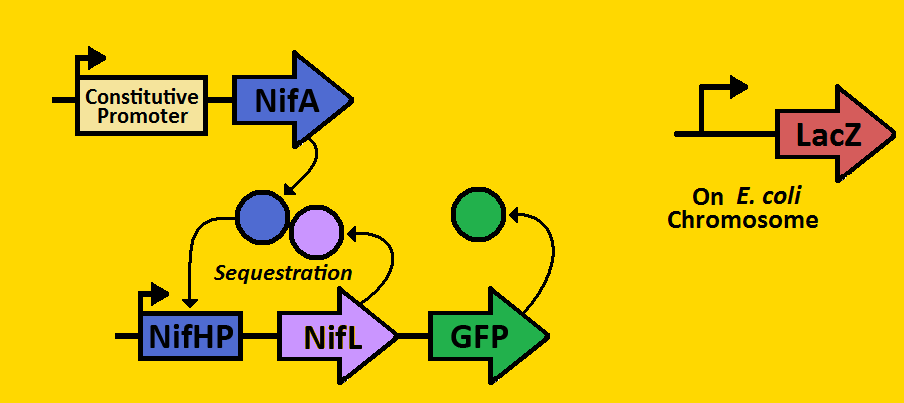
Project DescriptionCircadian ClocksShort background on circadian clocks... why they're important, why they're studied, maybe who studies them... Our Project: The SequestillatorWe can subdivide our clock into two parts: the activator module and the repressor module. The activator module consists of the constitutive promoter driving the NifA gene, thereby producing a constant amount of NifA. We will be using three different BioBrick promoters - corresponding to low, medium, and high outputs of NifA respectively. The NifA protein binds to the nifHp promoter of the repressor module, activating transcription of the NifL gene. Once NifL dimerizes, it can bind to the NifA hexamer, hence preventing NifA from binding to NifHp. This sequestration effect provides the clock's negative feedback loop that is essential for oscillations. Our hope is to put these modules on the E.coli chromosome using a BioBrick compatible Arabinose Landing Pad (our iGEM 2007 project) and a Leucine Landing Pad (a construct of a former Ninfa lab member, Dong Eun Chang). We will use E.coli strain NCM 1971, which has the nifHp driving lacZ on the chromosome. This way, we can test the amounts of NifA via Beta-galactosidase activity and test the amount of NifL via fluorescent microscopy. |
|---|
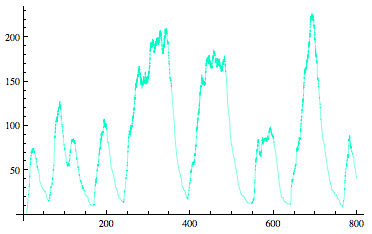
Sequestilator Modeling |
Sequestilator FabricationIf you like the way this looks, you could put a summary of what you built here and then we can have a separate page for fabrication, which might be a good idea. |
|---|
|
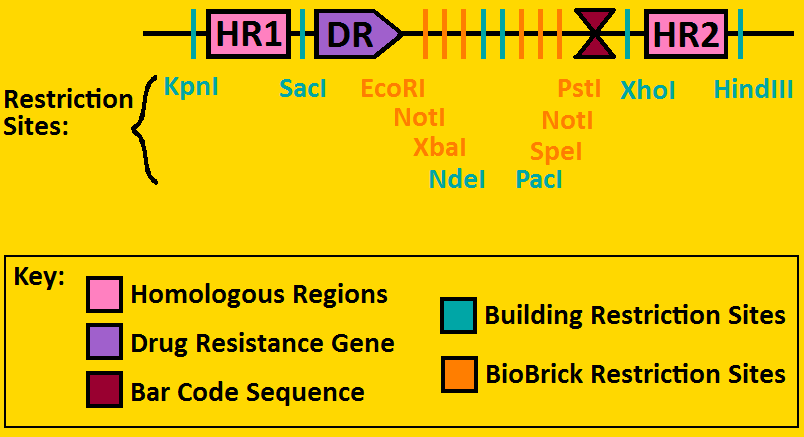
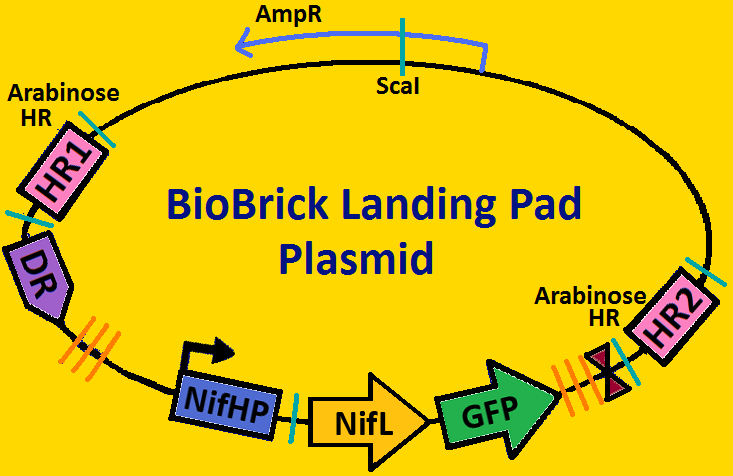
Landing PadsA landing pad is tool used to place synthetic operons on a chromosome. We will be using two landing pads for our project: the arabinose landing pad and leucine landing pad. Both of these plasmids will replace the respective metabolic operons with our desired subcloned genetic elements. The leucine landing pad was constructed by a former member of the Ninfa lab, Dong Eun Chang. The arabinose landing pad was a part of our iGEM 2007 project, and was worked on by Alyssa Delke and Khalid Miri. Our reason for using these pieces is to limit the noise in our system so that hopefully we can see more sustained oscillations than previous synthetic clocks have given.
Arabinose Landing Pad with our second operon inserted: [http://synthbio.engin.umich.edu/wiki/index.php/BioBrick_Landing_Pad Learn more about Landing Pads Here!]
|
|---|
 "
"