| Date and Time
| Gel check
| Protocol
| Comments & Actions
|
- 9/1, 08 PCR
- 9/2, 08 GEL check
|
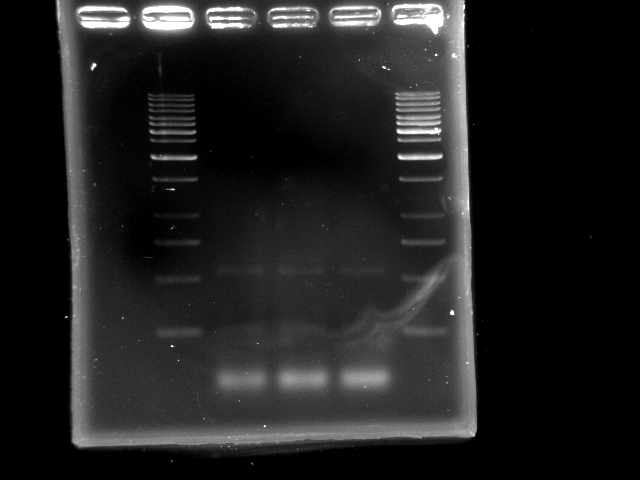
- lane 1: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 2: PCR pst tube #1
- lane 3: PCR pst tube #2
- lane 4: PCR pst tube #3
- lane 5: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1λ
- 10uM RP 1λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5λ
- PfuTubro 1λ (2.5U)
- ddH2O 38.5λ
- Reaction condition
- 95 10m, (95 30s, 55 1m, 72 3m15s) X 35, 72 5m
|
- Observations
- 3 nonspecific bands: ~500bp, 250bp and one less than 250bp (primer?)
- Does polymerase have sufficient time to elongate the 4.6kb pst operon?
- Actions
- Increase extension time from 3m15s to 5m
|
- 9/2, 08 PCR
- 9/3, 08 GEL check
|
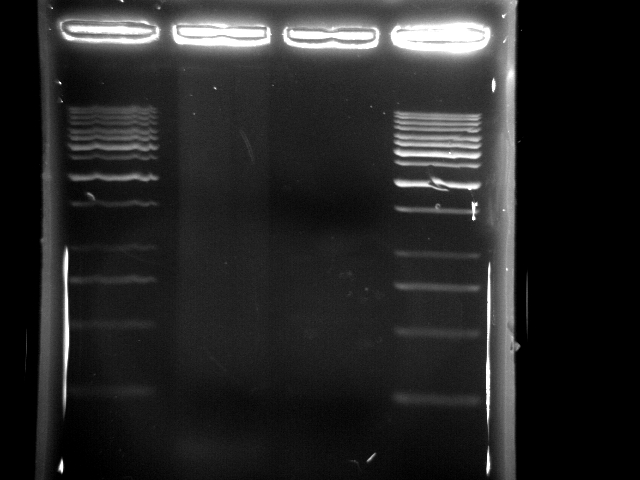
- lane 1: 1kb DNA ladder
- lane 2: PCR pst tube (PfuTubro)
- lane 3: PCR pst tube (YEA taq)
- lane 4: 1kb DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1λ
- 10uM RP 1λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5λ
- PfuTubro 1λ (2.5U) and YEA taq
- ddH2O 38.5λ
- Reaction condition
- 95 10m, (95 30s, 55 1m, 72 5m) X 30, 72 5m
|
- Observations
- taq generated a lot of non-specific DNA fragments and makes the whole lane smeared
- the lane using pfu was not clear in running gel
- Actions
- Increase extension time from 5m to 10m
- Increase pfu from 1uL to 2uL
|
- 9/3, 08 PCR
- 9/4, 08 GEL check
|
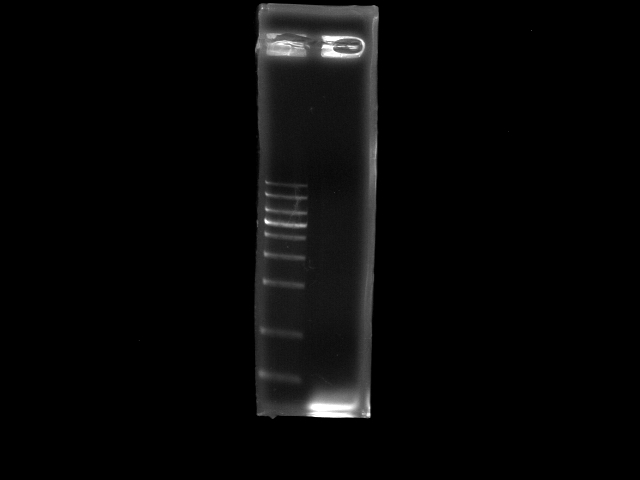
- lane 1: 1kb DNA ladder
- lane 2: PCR pst (double PfuTubro)
|
- Ingredients (Total 62λ)
- 50ng/λ template 5λ
- 10uM FP 20λ
- 10uM RP 20λ
- 2.5mM dNTP 5.0λ
- 10X buffer 9.17λ
- PfuTubro 2λ (5.0U)
- ddH2O 0.0λ
- Reaction condition
- 95 10m, (95 30s, 55 1m, 72 10m) X 30, 72 5m
|
- require a positive control (i.e. 3kb PCR from jesse or 2.6kb frmo tina)
- gradient Ta (55 - 65) to find the optimal one
|
- 9/5, 08 run PCR
- 9/5, 08 GEL check
|
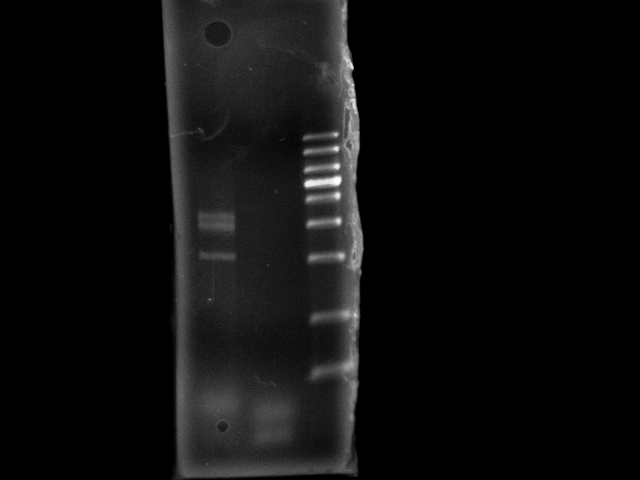
- lane 1: E0240 GFP (+ control, 1114bp)
- lane 2: pst
- lane 3: 1Kb DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1.0λ
- 10uM RP 1.0λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5.0λ
- YEA taq 1λ (5.0U)
- ddH2O 38.5λ
- Reaction condition
- 95 10m, (95 30s, 55 1m, 72 10m) X 30, 72 5m
|
- 10m extension time seems not sufficient to generate 4.6kb PST
|
|
|
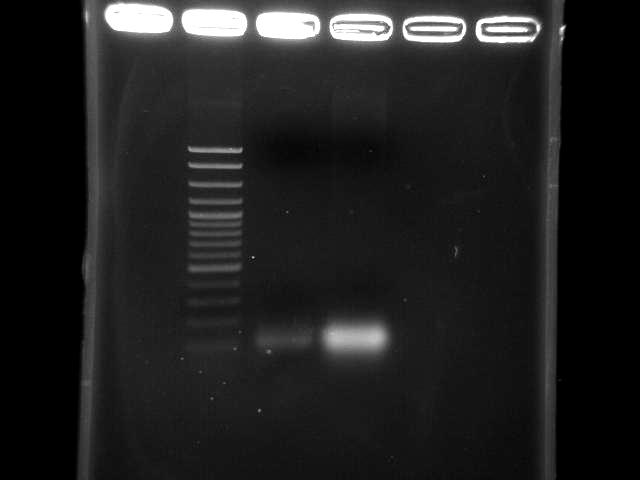
- lane 1: 100bp DNA ladder
- lane 2: pst by taq, 10m extension time (9/5 pst PCR)
- lane 3: pst by double TubroPfu, 10m extension time (9/3 pst PCR)
|
|
- two bands might be primers or short DNA fragments (~100bp)
- adjust reaction condition
- extension time 10m → 15m?
- annealing temperature 50 ~ 60
|
- 9/5, 9/6 PCR
- 9/6 GEL check
|
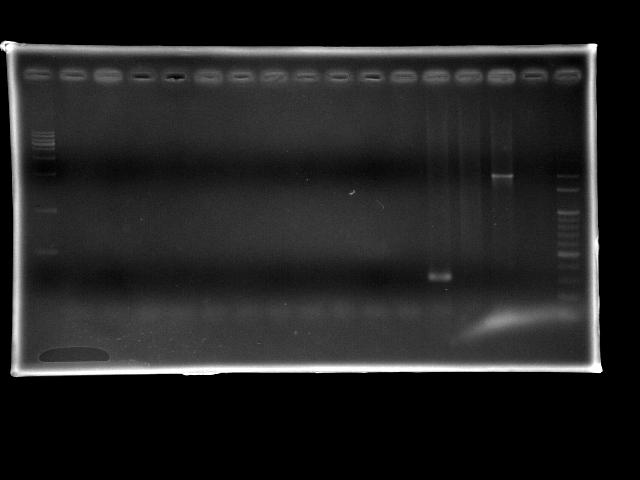
- lane 02: pst (Ta = 50.0°C, Textension = 15m)
- lane 03: pst (Ta = 50.2°C, Textension = 15m)
- lane 04: pst (Ta = 50.9°C, Textension = 15m)
- lane 05: pst (Ta = 52.0°C, Textension = 15m)
- lane 06: pst (Ta = 53.2°C, Textension = 15m)
- lane 07: pst (Ta = 54.4°C, Textension = 15m)
- lane 08: pst (Ta = 55.6°C, Textension = 15m)
- lane 09: pst (Ta = 56.8°C, Textension = 15m)
- lane 10: pst (Ta = 58.0°C, Textension = 15m)
- lane 11: pst (Ta = 59.1°C, Textension = 15m)
- lane 12: pst (Ta = 59.8°C, Textension = 15m)
- lane 13: ppk1 (Ta = 55°C, Textension = 6m)
- lane 14: ppk2 (Ta = 55°C, Textension = 6m)
- lane 15: htlB (Ta = 55°C, Textension = 6m)
- lane 16: Dye only (empty)
- lane 17: 100bp DNA ladder
|
PST
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1.0λ
- 10uM RP 1.0λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5.0λ
- YEA taq 1λ (5.0U)
- ddH2O 38.5λ
- Reaction condition
- 95 10m, (95 30s, 55 1m, 72 15m) X 30, 72 5m
|
- Observations
- pst PCR failed
- extension time is too long so that the taq polymerases were all dead before they can amplify enough amount of DNA fragments
- gradient search of Ta did not work
- ppk1 PCR works
- ppk2 PCR failed
- htlB PCR works (positive control)
|
|
|
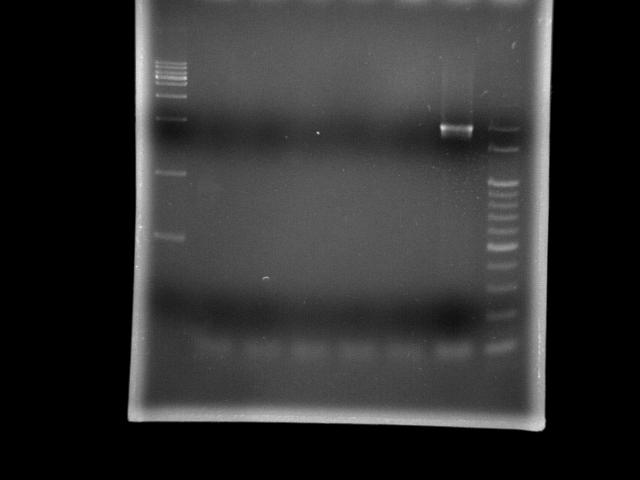
- lane 1: 1kb DNA ladder
- lane 2: pst (Ta = 45.0°C, Textension = 5m)
- lane 3: pst (Ta = 47.0°C, Textension = 5m)
- lane 4: pst (Ta = 49.4°C, Textension = 5m)
- lane 5: pst (Ta = 51.8°C, Textension = 5m)
- lane 6: pst (Ta = 55.0°C, Textension = 5m)
- lane 7: htlB (Ta = 55.0°C, Textension = 5m)
- lane 8: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1λ
- 10uM RP 1λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5λ
- YEA taq 1λ (5.0 Unit)
- ddH2O 38.5λ
- Reaction condition
- 95 2m30s, (95 30s, 55 30s, 72 5m) X 35, 72 5m
|
- Observation
- 95 2m30s is sufficient to open genomic DNA of E.coli K12
|
- 9/9, 08 PCR
- 9/9, 08 GEL check
|
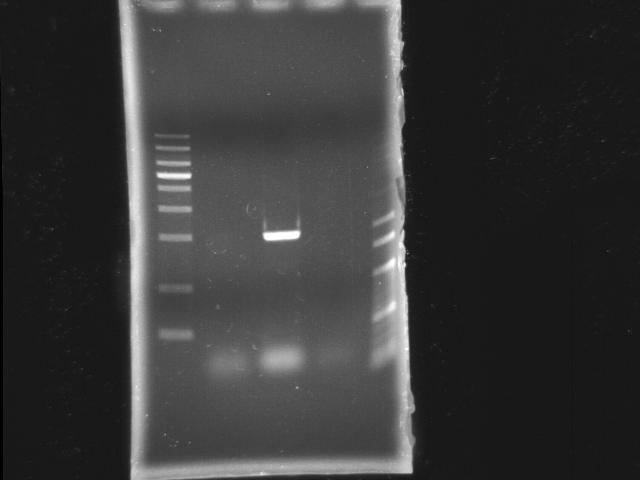
- lane 1: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 2: PCR pst
- lane 3: PCR htlB
- lane 4: PCR ppk2
- lane 5: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 2.5λ
- 10uM RP 2.5λ
- 2.5mM dNTP 4.0λ
- 10X buffer 5λ
- Promega pfu 1λ (1.5U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 55 30s, 72 9m30s) X 20, 72 5m
|
|
- 9/17, 08 Digestion
- 9/17, 08 GEL check
|
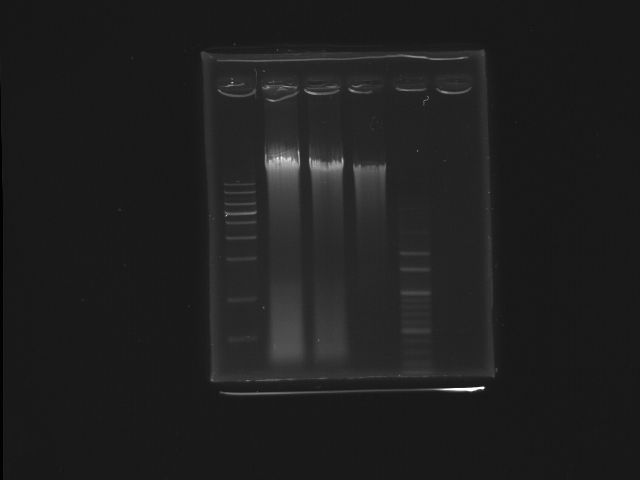
- lane01: 1Kb DNA marker
- lane02: K12 (SmaI digested)
- lane03: K12 (SmaI digested)
- lane04: K12 (oirginal)
- lane05: 100bp DNA marker
|
- Ingredients (Total λ)
- Stock concerntration of K12 genomic DNA:1.77mg/λ
- template:8λ
- NEBbuffer4:2λ
- BSA: 2λ
- 10units SmaI:1.416λ (1.77*8=10*X; X=1.416)
- Total reaction volume=20λ
- After digestion, we extracted the fragment(~7K)contained pst operon from gel for the use of template
|
- Motivation
- digest K12 genome DNA into smaller piece containing PST operon
|
|
|
|
|
|
- 9/17, 08 PCR
- 9/18, 08 GEL check
|
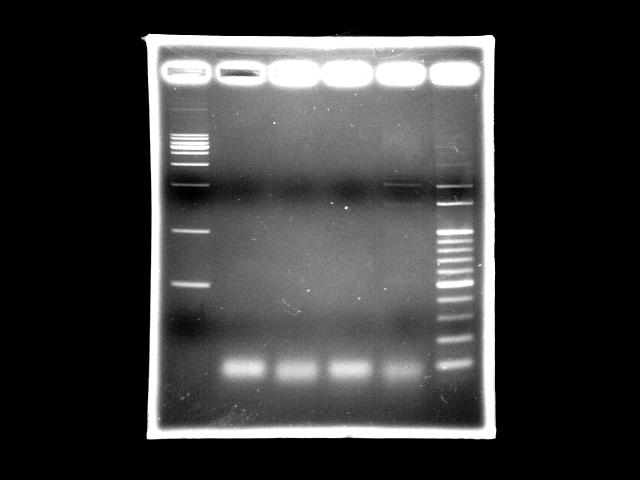
- lane 1: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 2: PPK overlapped PCR
- lane 3: PPK overlapped PCR
- lane 4: PST (template: SmaI digested K12 genomic DNA)
- lane 5: PST (template: original K12 genomic DNA)
- lane 6: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 3.62ng/λ SmaI digested template 1λ // 50ng/λ template 1λ
- 10uM FP 2.5λ
- 10uM RP 2.5λ
- 2.5mM dNTP 4.0λ
- 10X buffer 5λ
- 25mM MgSO4 10λ)
- Promega turbo 1λ (1.5U)
- ddH2O 24.0λ
- Reaction condition
- 95 2m, (95 30s, 51 60s, 72 9m30s) X 20, 72 5m
|
|
- 9/18, 08 PCR
- 9/19, 08 GEL check
|
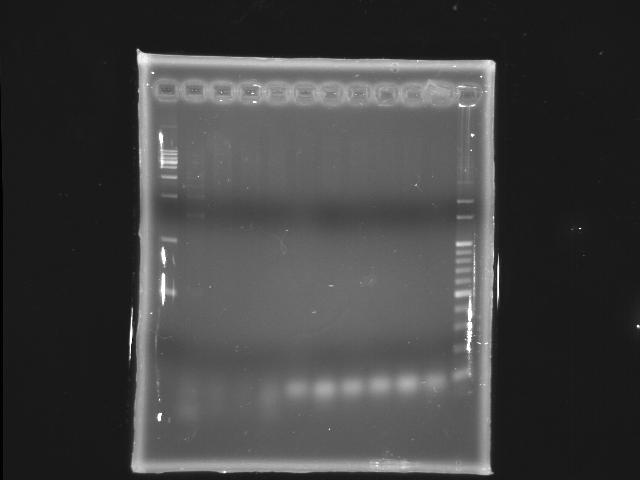
- lane 1: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 2: 45°C (with Mg2+)
- lane 3: 47°C (with Mg2+)
- lane 4: 48.2°C (with Mg2+)
- lane 5: 50.6°C (with Mg2+)
- lane 6: 53°C (with Mg2+)
- lane 7: 45°C
- lane 8: 47°C
- lane 9: 48.2°C
- lane 10: 50.6°C
- lane 11: 53°C
- lane12:100bp
|
- Ingredients (Total 25λ)
- 50ng/λ template 0.5λ
- 10uM FP 1.25λ
- 10uM RP 1.25λ
- 2.5mM dNTP 2λ
- 10X buffer 2.5λ
- 25mM MgSO4 5λ)
- Promega pfu 0.5λ (1.5U)
- ddH2O 12.0λ
- Reaction condition
- 95 2m, (95 30s, 45 47 48.2 50.6 53 60s, 72 9m30s) X 20, 72 5m
|
|
- 9/30, 08 gradient PCR
- 10/1, 08 GEL check
|
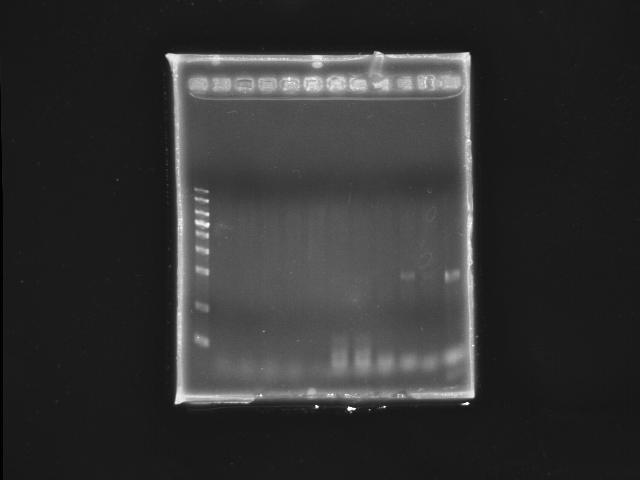
- lane 01: YEA 1Kb DNA ladder
- lane 02: PST right (black 1)
- lane 03: PST right (black 2)
- lane 04: PST right (black 3)
- lane 05: PST right (black 4)
- lane 06: PST right (black 5)
- lane 07: PST left (red 1)
- lane 08: PST left (red 2)
- lane 09: PST left (red 3)
- lane 10: PST left (red 4)
- lane 11: PST left (red 5)
|
- Ingredients (Total 25λ)
- 50ng/λ template ?λ
- 10uM FP 0.5λ
- 10uM RP 0.5λ
- 2.5mM dNTP 1.0λ
- 10X buffer 2.5λ
- YEA taq 0.125λ
- ddH2O 20.375λ
- Reaction condition
- 95 2m, (95 15s, 45-55 30s, 72 3m) X 30, 72 5m
|
- Observations
- no PST right segment can be amplified
- only one PST left segment was amplified under Ta =~ 53.0
|
- 10/1, 08 gradient PCR
- 10/1, 08 GEL check
|
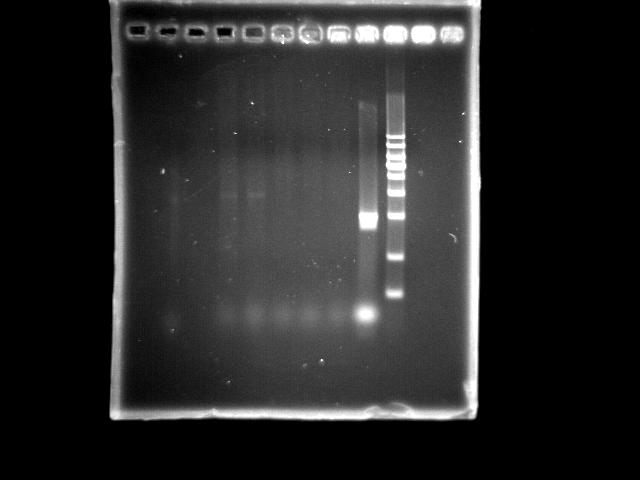
- lane 04: PST right (Ta = 45.0)
- lane 05: PST right (Ta = 47.0)
- lane 06: PST right (Ta = 50.6)
- lane 07: PST right (Ta = 53.0)
- lane 08: PST right (Ta = 55.0)
- lane 09: HtlB (+ control)
- lane 10: YEA 1Kb DNA ladder
|
- Ingredients (Total 25λ)
- 50ng/λ template ?λ
- 10uM FP 0.5λ
- 10uM RP 0.5λ
- 2.5mM dNTP 1.0λ
- 10X buffer 2.5λ
- YEA taq 0.125λ
- ddH2O 20.375λ
- Reaction condition
- 95 2m, (95 15s, 45;47;50.6;53;55 30s, 72 4m) X 30, 72 5m
|
- Observations
- Two PST right segments were amplified under Ta = 45.0 and 47.0
|
- 10/5, 08 fusion PCR
- 10/5, 08 GEL check
|
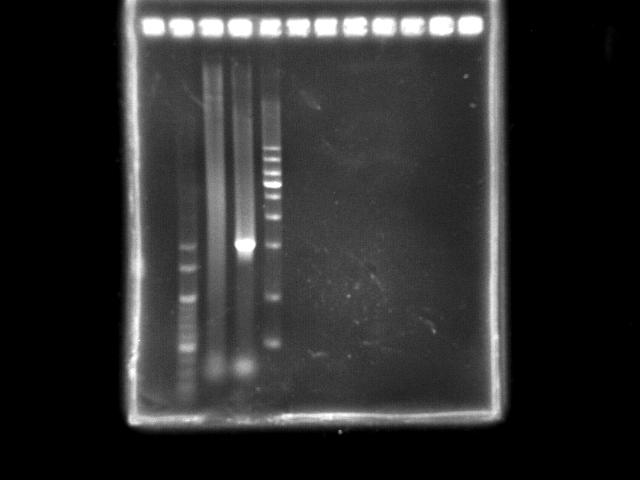
- lane 02: YEA 1Kb DNA ladder
- lane 03: PST fusion (Ta = 45.0)
- lane 04: HtlB (+ control)
- lane 05: YEA 1Kb DNA ladder
|
- Ingredients (Total 25λ)
- 50ng/λ template ?λ
- 10uM FP 0.5λ
- 10uM RP 0.5λ
- 2.5mM dNTP 1.0λ
- 10X buffer 2.5λ
- YEA taq 0.125λ
- ddH2O 20.375λ
- Reaction condition
- 95 2m, (95 30s, 45 60s, 72 8m) X 30, 72 5m
|
- Observations
- Combining two segments of PST operon was not successful (smeared). However, there was a bright region near the expected length (4.5Kb).
|
| Date and Time
| Gel check
| Protocol
| Comments & Actions
|
- 9/2, 08 PCR
- 9/3, 08 GEL check
|
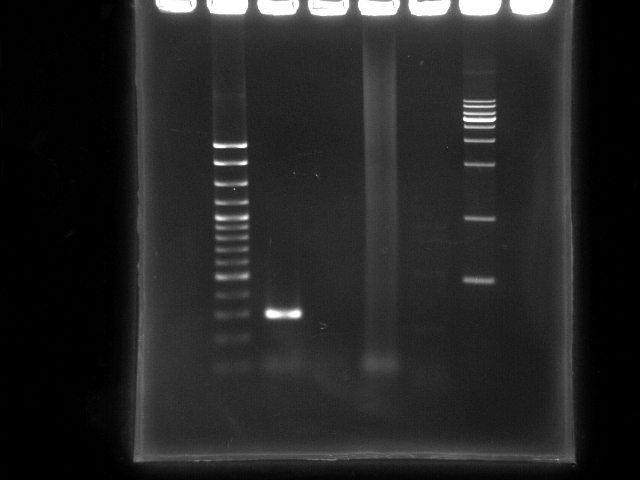
- lane 1: 100bp DNA ladder
- lane 2: PCR ppk 1 (left segment, ~ 270bp)
- lane 3: PCR ppk 2 (right segment, ~ 1797bp)
- lane 4: PCR pst (YEA taq)
- lane 5: PCR pst (PfuTubro)
- lane 6: 1kb DNA ladder
|
- ppk left segment(~ 270bp)
- 95 10m, (95 30s, 55 60s, 72 1m) x 35, 72 5m
- Promega pfu
- ppk right segment (~ 1797bp)
- 95 10m, (95 30s, 55 60s, 72 2m) x 35, 72 5m
- Promega pfu
- pst (~ 4.6kb)
|
- Observations
- PCR ppk1 has a clear band at ~300bp
- PCR ppk2 does not have any band on this lane
- PCR pst by taq clearly has a smear lane (a lot of non-specific annealing)
- PCR pst by TubroPfu has
- Actions
- increse extension time of ppk 2 (right segment)
- Notes
- ppk 1 (left segment) was lost >_<
|
- 9/5, 08 PCR
- 9/5, 08 GEL check
|

|
- ppk right segment (~ 1797bp)
- 95 10m, (95 30s, 55 60s, 72 3.5m) x 30, 72 5m
- Promega pfu
|
- lacking of positive control
- longer extension time(~4min)
|
- 9/9, 08 PCR
- 9/9, 08 GEL check
|

- lane 01: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 02: htlB (taq)
- lane 03: htlB (pfu)
- lane 04: PPK2 (taq)
- lane 05: PPK2 (pfu)
- lane 06: 100bp DNA ladder
|
- Ingradients(Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 2.5λ
- 10uM RP 2.5λ
- 2.5mM dNTP 4.0λ
- 10X buffer 5λ
- Promega pfu 1λ (1.5U)
- ddH2O 34.0λ
- Reaction condition
- 95 1'30m, (95 30s, 51 60s, 72 5m) X 20, 72 5m
- Suddently stop at the 13th cycle
|
- Observation
- though the PCR machine suddenly stop at the 13th cycle (Sensor Error), the positive control HtlB with Taq polymerase still have a band at 2Kb.
- Action
- We moved the PCR product tube to the other PCR machine, continuing the remained 7 cycles.
|
- 9/9, 9/10 08 PCR
- 9/10, 08 GEL check
|
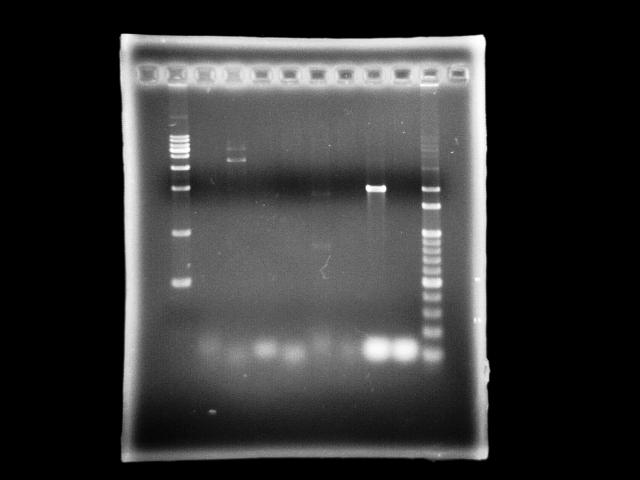
- lane 01: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
- lane 2: 9/10, ppk2 (taq)
- lane 3: 9/10, ppk2 (pfu)
- lane 4: 9/10, htlB (taq)
- lane 5: 9/10, htlB (pfu)
- lane 6: 9/9, ppk2 (taq)
- lane 7: 9/9, ppk2 (pfu)
- lane 8: 9/9, htlB (taq)
- lane 9: 9/9, htlB (pfu)
- lane 10: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 2.5λ
- 10uM RP 2.5λ
- 2.5mM dNTP 4.0λ
- 10X buffer 5λ
- Promega pfu 1λ (1.5U)
- ddH2O 34.0λ
- Ingredients (Total 50λ)
- 50ng/λ template 1λ
- 10uM FP 1.0λ
- 10uM RP 1.0λ
- 2.5mM dNTP 2.5λ
- 10X buffer 5λ
- YEA taq 1λ (5.0U)
- ddH2O 38.5λ
- Reaction condition (9/9, 08)
- 95 2m, (95 30s, 51 60s, 72 5m) X 20, 72 5m
- Reaction condition (9/10, 08)
- 95 2m, (95 30s, 50 60s, 72 9m30s) X 20, 72 5m
|
- Observations
- PPK2 has ~1.8kb band by taq (Ta = 51, te = 5m)
- assume annealing problem is universal, we use taq to find a optimal Ta
- in case we find it, we can apply this Ta to pfu PCR
- Actions
- use Ta gradient to find the optimal Ta
|
- 9/10, 08 PCR
- 9/10, 08 GEL check
|
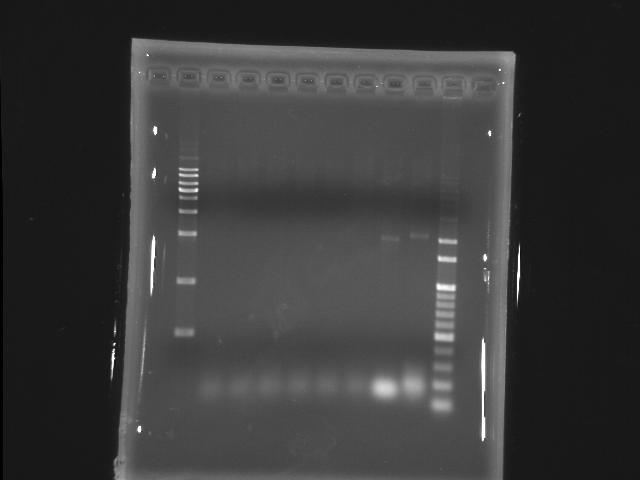
- lane 01: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
PPK2
- lane 02: 50.9
- lane 03: 53.2
- lane 04: 55.6
- lane 05: 56.8
- lane 06: 59.1
- lane 07: 60.0
- lane 10: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- NEB taq 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, [51-59:2] 60s, 72 2m) X 20, 72 5m
|
- Actions
- longer extension time (2m to 5m)
- increase the amount of cycles (20 to 25)
|
- 9/10, 08 PCR
- 9/11, 08 GEL check
|
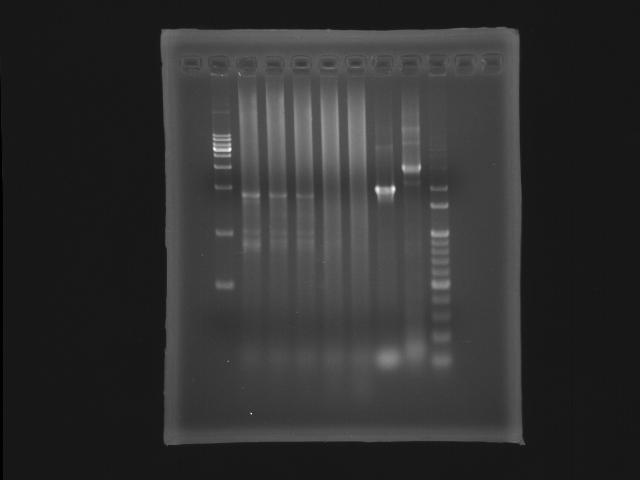
- lane 01: [http://www.fermentas.com/catalog/electrophoresis/generulers.htm#1kb 1kb DNA ladder (fermentas)]
PPK2
- lane 02: 47.6
- lane 03: 51.4
- lane 04: 52.8
- lane 05: 54.0
- lane 06: 56.0
- lane 09: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- NEB taq 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 1m, (95 30s, [47.6-56:2] 60s, 72 5m) X 25, 72 5m
|
- Observations
- we got the correct band (~1.8K) at Ta=52.8,51.4 and 47.6 degree celcius but also find that the PPK2 lanes smear serously.
- Actions
- substituting Taq ploymerase to Promega (pfu)
- longer the extension time (5m to 6m30s)
|
- 9/11, 08 PCR
- 9/11, 08 GEL check
|
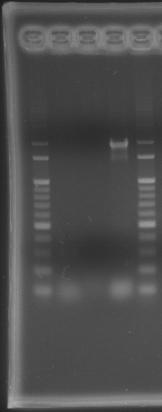
- lane 01: 100bp DNA ladder
- lane 02: PPK1
- lane 03: PPK2
- lane 05: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- Promega pfu 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 53 60s, 72 6'30m) X 25, 72 5m
|
- Observations
- there were no band on the gel, PCR condition failure.
|
- 9/11, 08 PCR
- 9/14, 08 GEL check
|
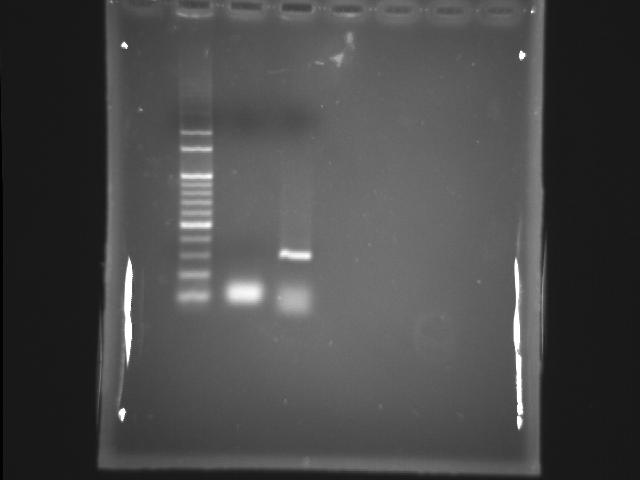
- lane 01: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- Promega pfu 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 55 60s, 72 1m) X 20, 72 5m
|
- Observations
- Find the correct and clear PPK1 band at ~273bp
- Actions
- Store the correct PCR Product in the 1.2ml eppendorf at -20 degree celcius refrigerator
|
- 9/15, 08 PCR
- 9/15, 08 GEL check
|
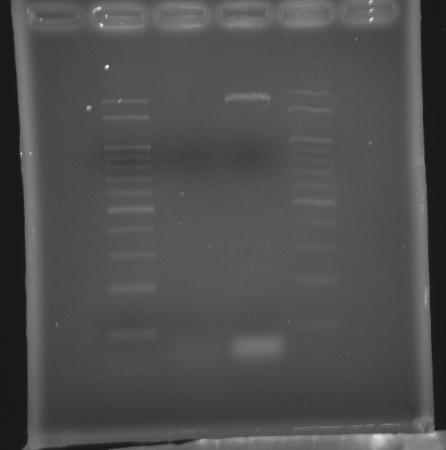
- lane 01: 100bp DNA ladder
- lane 04: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- Promega pfu 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 51 60s, 72 6m30s) X 20, 72 5m
|
- Observations
- No band appears in lane 02
- Actions
- We had longer but incorrect band using promega pfu (Ta = 50 and extension time = 9m30s)
- Increase extension time from 6m30s to 9m30s
|
- 9/15, 08 PCR
- 9/16, 08 GEL check
|

- lane 02: Pst operon
- lane 03: PPK2
- lane 04: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- pfu turbo 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 51 60s, 72 9m30s) X 20, 72 5m
|
- Actions
- We re-run the remaining PPK2 PCR product to make the band more clear (the DNA concentration got higher) in order to do gel extraction of specific band (1.7K)
- Run nested PCR to amplify the PPK2 target DNA fragment
|
- 9/16, 08 nested PCR
- 9/17, 08 GEL check
|
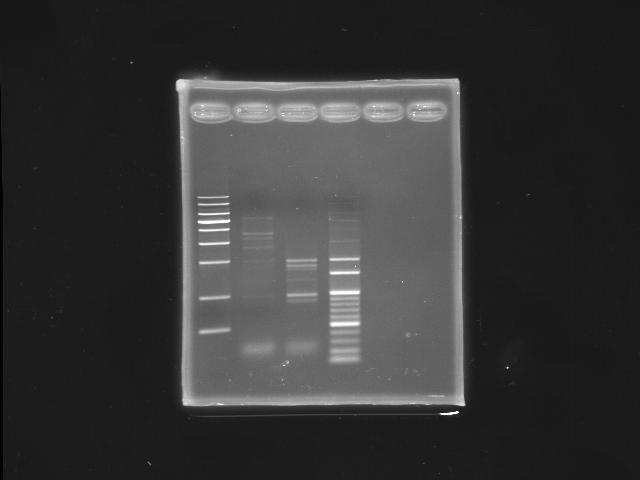
- lane 02: PPK2 (K12 as template)
- lane 03: PPK2 (PPK2 as template, nested PCR)
- lane 04: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 1λ (1ng/λ = 1ug/mL)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- pfu turbo 1λ (5.0U)
- ddH2O 34.0λ
- Reaction condition
- 95 2m, (95 30s, 51 60s, 72 9m30s) X 20, 72 5m
|
|
- 9/20, 08 PCR
- 9/21, 08 GEL check
|
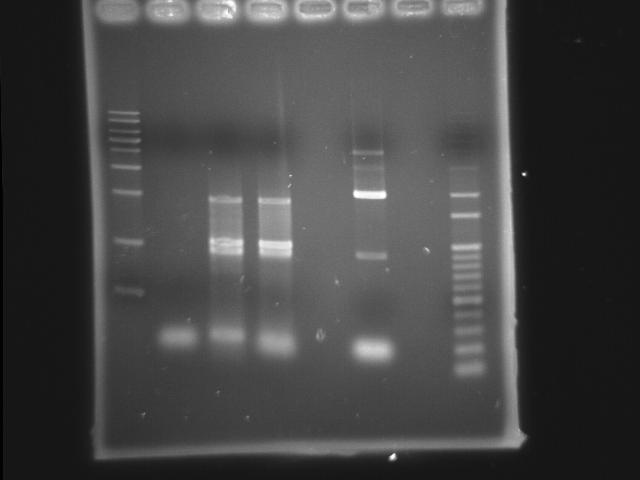
- lane 02: Pst (9/18 gradient-45)
- lane 03: PPK overlapped(Mg2+) (PPK1(9/11) PPK2 III(9/20))
- lane 04: PPK overlapped (PPK1(9/11) PPK2 III(9/20))
- lane 07: 100bp DNA ladder
|
- Ingredients (Total 50λ)
- 50ng/λ template 2λ (PPK1(9/11)1λ PPK2 III(9/20)1λ)
- 10uM FP 2.5λ (0.5uM)
- 10uM RP 2.5λ (0.5uM)
- 2.5mM dNTP 4.0λ (200uM)
- 10X buffer 5λ
- pfu promega 1λ (5.0U)
- ddH2O 33.0λ
- Reaction condition
- 95 2m, (95 30s, 51 60s, 72 9m30s) X 20, 72 5m
|
- Observations
- PST (Ta=45) band disappeared
- around 2kb has a band in PPK overlapped PCR
|
|
|
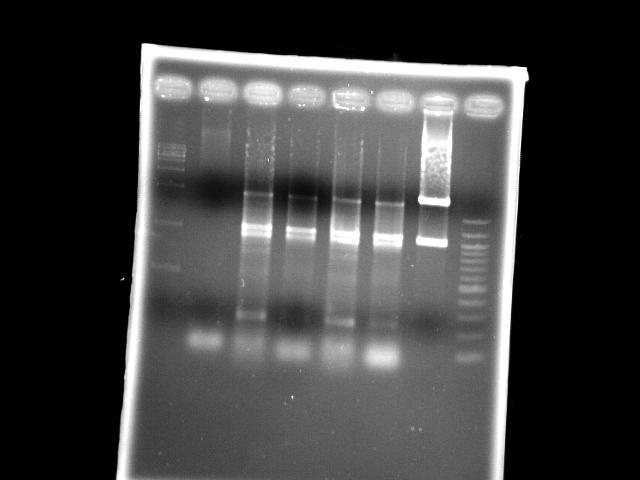
- lane 03: PPK (Mg2+, EcoRI digested)
- lane 04: PPK (EcoRI digested)
- lane 05: PPK (Mg2+)
- lane 06: PPK
- lane 08: NEB 100bp DNA ladder (1517, 1200, 1000, ...)
|
|
- Observations
- EcoRI digestion does not change the band pattern of PPK PCR product
- If EcoRI works, these PPK should be mutants
|
|
|
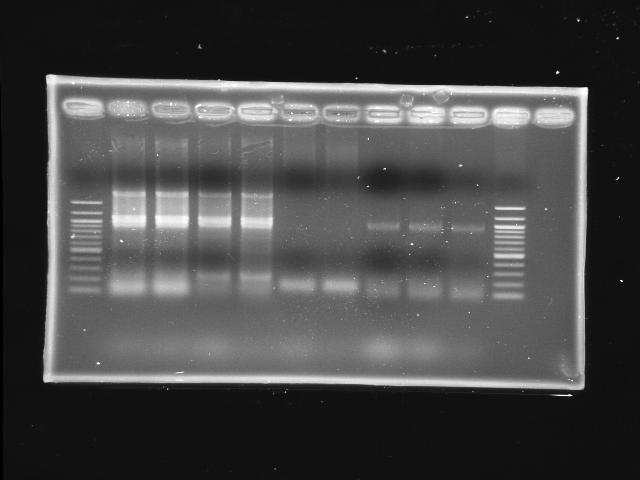
- lane 01: NEB 100bp DNA ladder (1517, 1200, 1000, ...)
- lane 2,3,4,5: PPK (pfu overlapped)
- lane 6,7: pst (KOD)
|
|
- Observations
- PPK indeed has a band around 2kb at lane 2-5 (cut it as insert)
- PST does not have any band (still hanging >_<)
|
| Date and Time
| Gel check
| Protocol
| Comments & Actions
|
- 9/19, 08 colony PCR
- 9/20, 08 GEL check
|
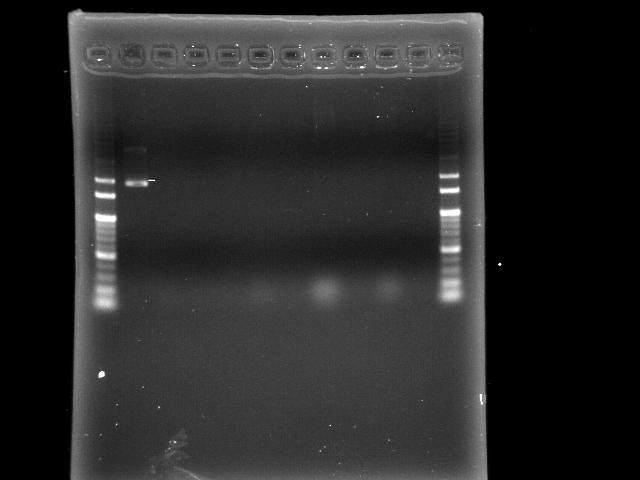
- lane 01: 100bp
- lane 02: positive control (E0240?)
- lane 03 - 10: P0440 colony #1 - #8
- lane 11: negative control (no template)
- lane 12: 100bp
|
- Reaction condition
- 95 2m, (95 30s, 55 30s, 72 60s) X 25, 72 7m
- we ran less cycles than usual (30 cycles)
|
- Observations
- no band at lane 03 - 10 (expected VRVF2 length: 1078)
- positive control is not correct
- Actions
- increase the number of cycles to 30
|
- 9/20, 08 colony PCR
- 9/20, 08 GEL check
|
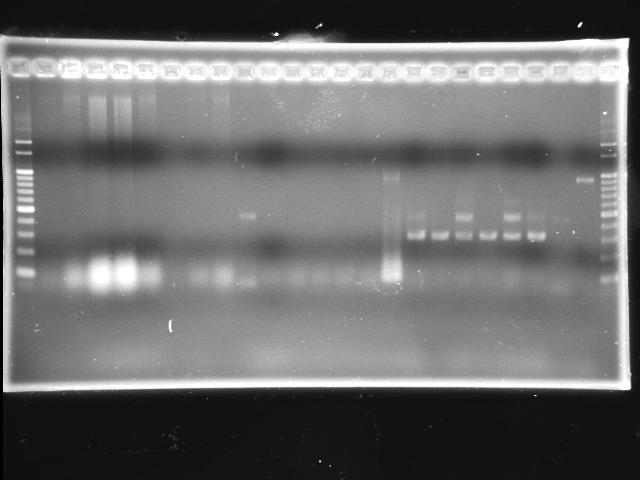
- lane 01: 100bp marker
- lane 02 - 09: P0440 colony #1 - #8
- lane 10: J04430-07 colony #1
- lane 11: I763004-08 colony #1
- lane 12: J04430-08 colony #1
- lane 12 - 14: J04430-07 colony #2 - #4
|
|
- Observations
- P0440 (expected VRVF2 length: 1078): no hit; one 500bp band is not correct
- J04430 (expected VRVF2 length: 1321): no hit
- I763004 (expected VRVF2 length: 1360): not hit
- colony PCR protocol may work
- Actions
- pick more colony by the same protocol
|
- 9/21, 08 colony PCR
- 9/21, 08 GEL check
|

- lane 01: 100bp marker (forget to load >_<)
- lane 02 - 08: pLac + E0240 colony #1 - #3, #9 - #12
- lane 09 - 15: P0440 colony #9 - #15
- lane 16 - 22: J04430-07 colony #5 - #11
- lane 23: positive control (pPhoB + E0240 colony #2; around 1.6Kb)
- lane 24: negative control (pUC19)
- lane 25: 1kb marker (forget to load >_<)
|
|
- Observations
- no DNA marker loaded; use positive control (1.6Kb) as marker instead
- pLac + E0240 (expected VRVF2 length: 1320): hit ratio 7/7
- P0040 (expected VRVF2 length: 1078): hit ratio 7/7
- J04430 (expected VRVF2 length: 1321): hit ratio 5/7
|
- 9/26, 08 colony PCR
- 9/26, 08 GEL check
|
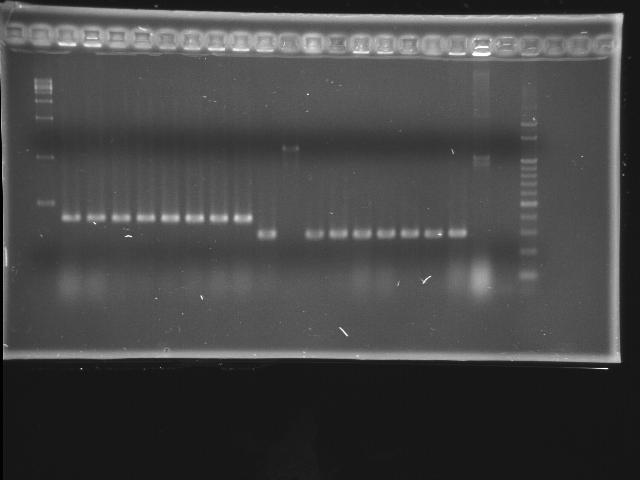
- lane 01: empty
- lane 02: 1kb marker
- lane 03 - 10: pLac + P0440 colony #1 - #8
- 1284 bp = 200 + 6 + 840 + 238
- lane 11 - 19: pTet + E0240 colony #1 - #8
- 1174 bp = 54 + 6 + 876 + 238, #7 repeats twice
- lane 20: positive control (P0440)
- lane 21: negative control (w/o template)
- lane 22: 100kb maker
- lane 23: empty
- lane 24: empty
- lane 25: empty
|
- Ingradients (0.5X)
- VR, VF2 1 uL
- 10X buffer 2.5 uL
- dNTP 1 uL
- taq 0.125 uL
- ddH2O 20.375 uL
- Reaction condition
- AB new PCR machine, protocol name: colony
|
- Observations
- pLac + P0440 (expected VRVF2 length: 1284): no hit
- pTet + E0240 (expected VRVF2 length: 1174): hit ratio 1/8
- colony #2 of pTet + E0240 has correct length
- Actions
- liquid culture colony #2 of pTet + E0240 and extract plasmid
|
- 9/27, 08 colony PCR
- 9/27, 08 GEL check
|
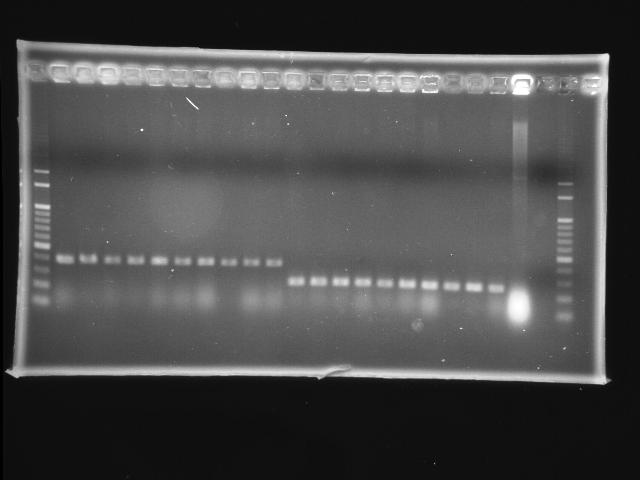
- lane 01: 100bp marker
- lane 02 - 11: pLac + P0440 (first ligation) colony #9 - #18
- lane 12 - 21: RBS + PPK colony #1 - #10
- lane 22: P0440 (positive control)
- lane 23: negative control (w/o template)
- lane 24: 100bp marker
- lane 25: empty
|
|
- Observations
- new ligation of pLac + P0440: no hit
|
- 9/27, 08 colony PCR
- 9/27, 08 GEL check
|
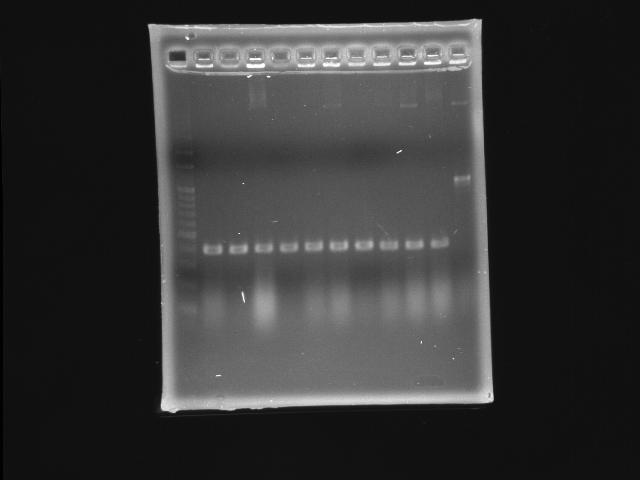
- lane 01: 100bp marker
- lane 02 - 11: pLac + P0440 (second ligation) colony #1 - #10
- lane 12: P0440 (positive control)
|
|
|
- 10/1, 08 colony PCR
- 10/1, 08 GEL check
|
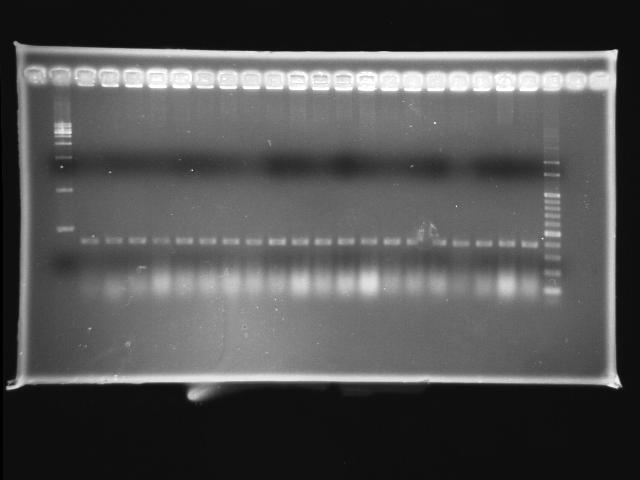
- lane 01: empty
- lane 02: 1kb marker
- lane 03 - 22: pLac + P0440 colony #19 - #38
- lane 23: 100kb maker
- lane 24: empty
- lane 25: empty
|
- Ingradients (0.5X)
- VR, VF2 1 uL
- 10X buffer 2.5 uL
- dNTP 1 uL
- taq 0.125 uL
- ddH2O 20.375 uL
|
- Observations
- pLac + P0440 (expected VRVF2 length: 1284): no hit
- Actions
- re-ligation of pLac + P0440
|
- 10/1, 08 colony PCR
- 10/1, 08 GEL check
|
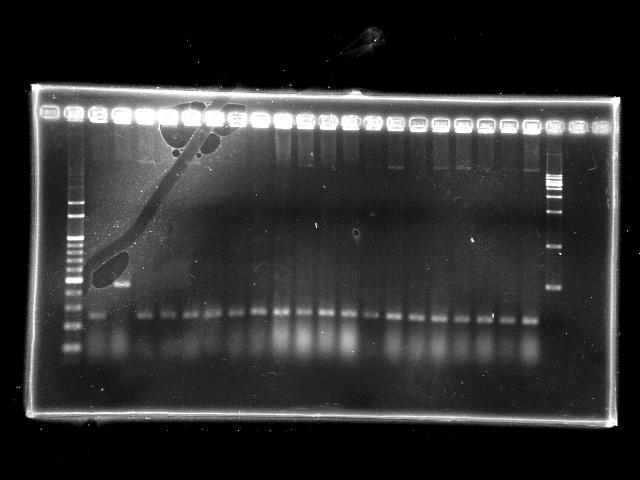
- lane 01: empty
- lane 02: 1kb marker
- lane 03 - 22: RBS + PPK colony #11 - #30
- lane 23: 100kb maker
- lane 24: empty
- lane 25: empty
|
- Ingradients (0.5X)
- VR, VF2 1 uL
- 10X buffer 2.5 uL
- dNTP 1 uL
- taq 0.125 uL
- ddH2O 20.375 uL
|
- Observations
- RBS + PPK (expected VRVF2 length: 2324): no hit
- Actions
- extension time (1m15s) is too short to amplify the 2.3Kb RBS + PPK
- increase the extension time to 2m20s
- However, the clear band at around 300bp indicates the ligation failed
|
 "
"




































