Team:NYMU-Taipei/Project/Time Regulation/Results of Cyanoxilator
From 2008.igem.org
Revision as of 00:26, 30 October 2008 by Blackrabbit (Talk | contribs)
| Home | Project Overview: | pH Sensor | Attachment | Time Regulation | Waste Removal | Experiments and Parts | About Us |
Contents |
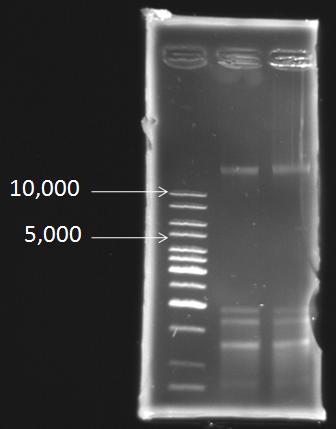
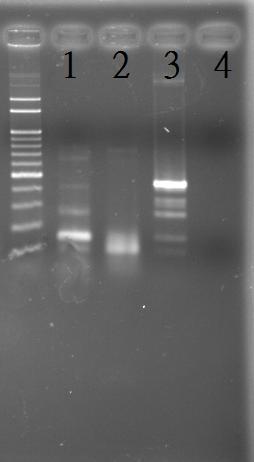
Extraction of the genomic DNA of S. elongatus PCC 7942
|
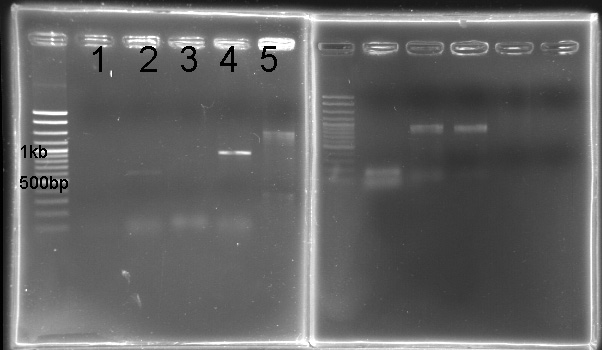
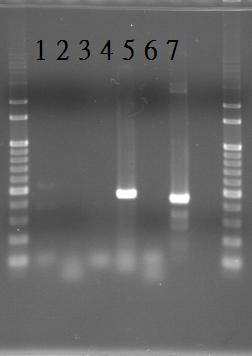
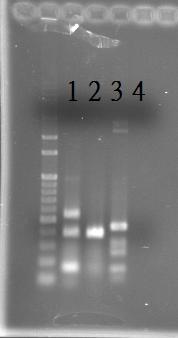
PCR of RpaA and SasA
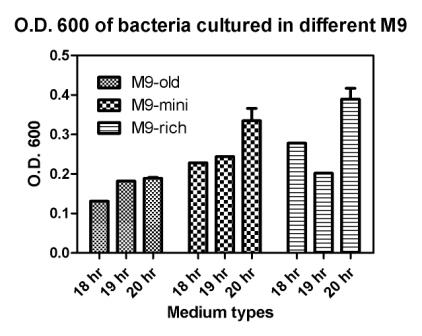
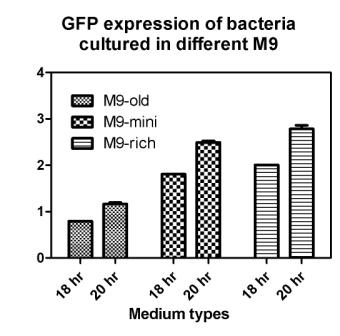
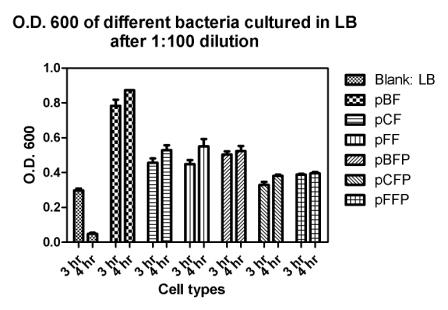
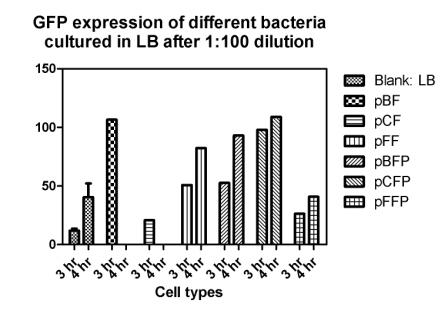
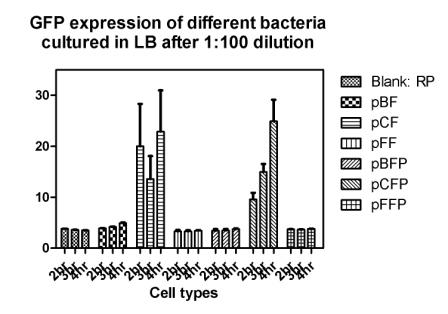
Reporting assay
Discussion of the reporting
- Are bacteria fresh or not important to GFP expression? If so, we should use freshly streaked plates.
- The white and black plates must clean carefully, otherwise, there would be too much background.
- pBF seem to grow much faster then others.
 "
"