Team:Newcastle University
From 2008.igem.org
| Line 16: | Line 16: | ||
''B. subtilis'' is our favorite bacteria to work with because it is gram positive, relatively safe to work with, is naturally competent, naturally sporulates, and is well-characterised for experimentation. | ''B. subtilis'' is our favorite bacteria to work with because it is gram positive, relatively safe to work with, is naturally competent, naturally sporulates, and is well-characterised for experimentation. | ||
| - | [[Team:Newcastle_University/Bacillus_subtilis|Learn more about B. subtilis]]. | + | [[Team:Newcastle_University/Bacillus_subtilis|Learn more about ''B. subtilis'']]. |
== Sponsors == | == Sponsors == | ||
{{:Team:Newcastle University/Sponsors}} | {{:Team:Newcastle University/Sponsors}} | ||
{{:Team:Newcastle University/HomeSideBar}} | {{:Team:Newcastle University/HomeSideBar}} | ||
Revision as of 15:50, 2 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Welcome! This year is Newcastle's first year participating in the iGEM competition! We are a small team of six students, three supervisors, and four advisors, but we have high ambitions for our first year. Recognizing the problem of antibiotic resistant bacteria and poor methods of diagnosis in underdeveloped countries, we aim to use synthetic biology and bioinformatics to engineer a simple, safe, fast, and reliable biological diagnostic system to identify bacteria.
Biological Theme: Bug Busters
We are developing a diagnostic tool that enables the detection of a range of Gram-positive bacterial pathogens endemic in patients around the world. This new system will enable visual detection of these pathogens within minutes. This application would have importance not only in hospital settings, but also in the third world.
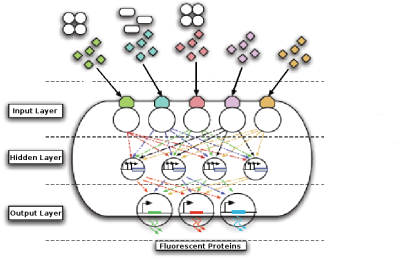
An important part of our approach is bioinformatics. We will produce a workbench that will incorporate a parts repository, constraints repository and an evolutionary algorithm (EA). The EA will take input from the parts repository and constraints repository to evolve a neural network simulation. The fittest model will be used to generate a DNA sequence which will implement the neural network in vivo. This DNA sequence will be synthesized and cloned into the Bacillus subtilis chassis. One of our outcomes will be a range of neural network node BioBrick devices which can be combined to form the in vivo neural network.
Bacillus subtilis
B. subtilis is our favorite bacteria to work with because it is gram positive, relatively safe to work with, is naturally competent, naturally sporulates, and is well-characterised for experimentation.
Sponsors

|

|

|

|

|
 "
"