Team:Paris/Modeling/Programs
From 2008.igem.org
(Difference between revisions)
(→Finding Parameters) |
|||
| Line 1: | Line 1: | ||
{{Paris/Menu}} | {{Paris/Menu}} | ||
| - | + | {|cellspacing="5" cellpadding="10" style="background:#649CD7; width: 965px;" | |
| + | |-valign="top" | ||
| + | |style="background:#ffffff"| | ||
==Finding Parameters== | ==Finding Parameters== | ||
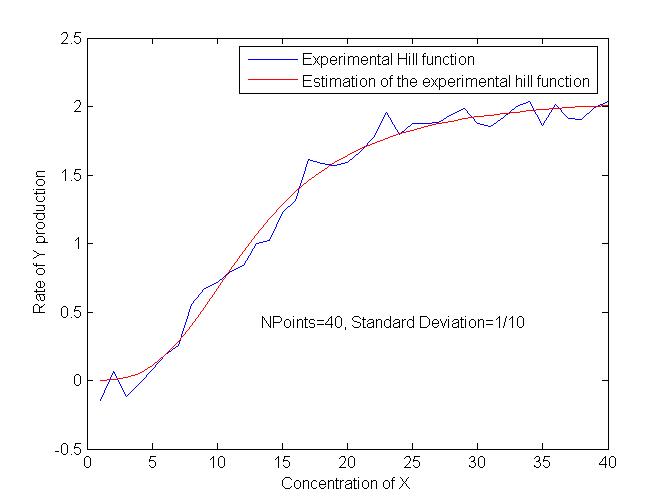
This program aims at fill [[Team:Paris/Modeling/Bibliography|this data bank]], from experimental data obtained by our wet lab. | This program aims at fill [[Team:Paris/Modeling/Bibliography|this data bank]], from experimental data obtained by our wet lab. | ||
| Line 18: | Line 20: | ||
After some trials, we have obtained interesting results in terms of precision. We are now trying to test the robustness of the estimations in order to quantify the influence of biological variance in the results as well as the influence of the number of points available. | After some trials, we have obtained interesting results in terms of precision. We are now trying to test the robustness of the estimations in order to quantify the influence of biological variance in the results as well as the influence of the number of points available. | ||
| - | [[Image:Estimation.jpg]] | + | [[Image:Estimation.jpg|center]] |
The corresponding code can be found there : [[Team:Paris/Modeling/|Estimation of the parameters]] | The corresponding code can be found there : [[Team:Paris/Modeling/|Estimation of the parameters]] | ||
| + | |||
| + | |}<br style="clear:both" /> | ||
Revision as of 13:55, 30 July 2008
|
 "
"