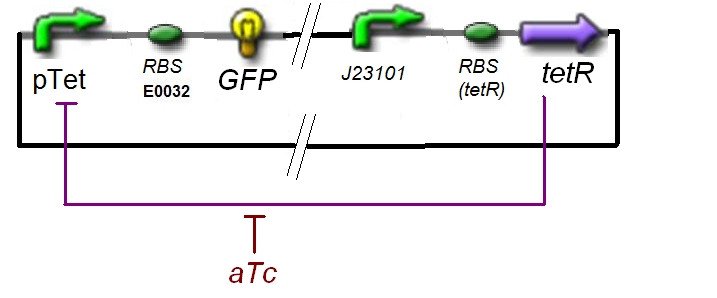
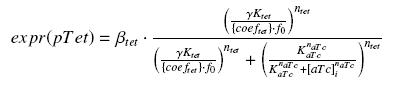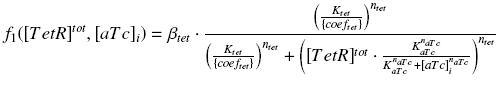find_ƒ1
function optimal_parameters = find_f1(X_data, Y_data, initial_parameters)
function output = expr_pTet(parameters, X_data)
for k = 1:length(X_data)
output(k) = parameters(1) * (1 - ...
hill((1 - hill(X_data(k),parameters(4),parameters(5))),parameters(2),parameters(3)));
end
end
options=optimset('LevenbergMarquardt','on','TolX',1e-10,'MaxFunEvals',1e10,'TolFun',1e-10,'MaxIter',1e4);
optimal_parameters = lsqcurvefit( @(parameters, X_data) expr_pTet(parameters, X_data), ...
initial_parameters, X_data, Y_data, 1/10*initial_parameters, 10*initial_parameters, options );
end
Inv_ƒ1
function quant_aTc = Inv_f1(inducer_quantity)
global gamma, f0;
function equa = F(x)
equa = f1( (f0/gamma) , x ) - inducer_quantity;
end
options=optimset('LevenbergMarquardt','on','TolX',1e-10,'MaxFunEvals',1e10,'TolFun',1e-10,'MaxIter',1e4);
quant_aTc = fsolve(F,1,options);
end
 "
"