Team:Paris/Modeling/f2
From 2008.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
|[expr(pBad)] | |[expr(pBad)] | ||
|expression rate of <br> pBad '''with RBS E0032''' | |expression rate of <br> pBad '''with RBS E0032''' | ||
| - | |nM. | + | |nM.min<sup>-1</sup> |
|see [[Team:Paris/Modeling/Programs|"findparam"]] <br> need for 20 measures | |see [[Team:Paris/Modeling/Programs|"findparam"]] <br> need for 20 measures | ||
|- | |- | ||
|γ<sub>GFP</sub> | |γ<sub>GFP</sub> | ||
|dilution-degradation rate <br> of GFP(mut3b) | |dilution-degradation rate <br> of GFP(mut3b) | ||
| - | | | + | |min<sup>-1</sup> |
| - | | | + | |0.0198 |
|- | |- | ||
|[GFP] | |[GFP] | ||
| Line 50: | Line 50: | ||
|β<sub>bad</sub> | |β<sub>bad</sub> | ||
|production rate of pBad '''with RBS E0032''' <br> not in the system | |production rate of pBad '''with RBS E0032''' <br> not in the system | ||
| - | |nM. | + | |nM.min<sup>-1</sup> |
| | | | ||
|- | |- | ||
| - | |(K<sub>bad</sub>/[AraC<sub>tot</sub>] | + | |(K<sub>bad</sub>/[AraC<sub>tot</sub>]) |
|activation constant of pBad <br> not in the system | |activation constant of pBad <br> not in the system | ||
| - | |nM | + | |nM |
| | | | ||
|- | |- | ||
| Line 65: | Line 65: | ||
|K<sub>ara</sub> | |K<sub>ara</sub> | ||
|complexation constant Arabinose-AraC <br> not in the system | |complexation constant Arabinose-AraC <br> not in the system | ||
| - | |nM | + | |nM |
| | | | ||
|- | |- | ||
Revision as of 22:18, 4 September 2008
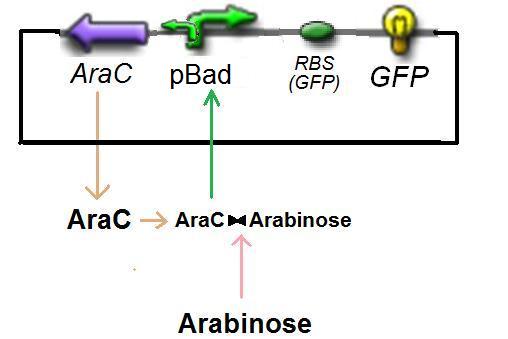
According to previous considerations on pBad, we have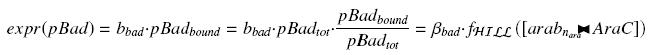
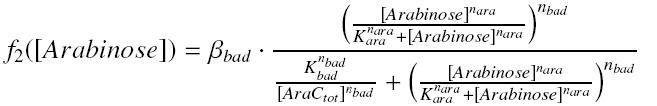
so, with the complexation between Arabinose and AraC, and after reducing the equation
| param | signification | unit | value |
| [expr(pBad)] | expression rate of pBad with RBS E0032 | nM.min-1 | see "findparam" need for 20 measures |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 |
| [GFP] | GFP concentration at steady-state | nM | need for 20 measures |
| (fluorescence) | value of the observed fluorescence | au | need for 20 measures |
| conversion | conversion ration between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value |
| βbad | production rate of pBad with RBS E0032 not in the system | nM.min-1 | |
| (Kbad/[AraCtot]) | activation constant of pBad not in the system | nM | |
| nbad | complexation order of pBad not in the system | no dimension | |
| Kara | complexation constant Arabinose-AraC not in the system | nM | |
| nara | complexation order Arabinose-AraC not in the system | no dimension |
 "
"