Team:Rice University/CONSTRUCTS
From 2008.igem.org
DavidOuyang (Talk | contribs) |
DavidOuyang (Talk | contribs) |
||
| Line 19: | Line 19: | ||
==='''Constructs'''=== | ==='''Constructs'''=== | ||
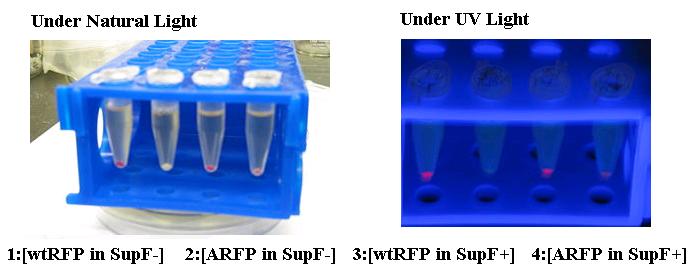
| - | <p>''Saccharomyces cerevisiae'' are widely used for baking and brewing | + | <p>''Saccharomyces cerevisiae'' are widely used for baking and brewing are particularly useful for synthesizing metabolites under fermentation conditions. Such microaerobic conditions prevent the air oxidation of bioreactive compounds and are optimal for the ''in vivo'' synthesis of resveratrol. To achieve our project and expand the synthetic biology toolbox for programming yeast, we have introduced into the iGem registry BioBricks encoding 3 yeast promoters, 3 yeast terminators, a two micron origin of replication, 2 selectable markers, 2 enzymes, and a yeast integration plasmid. In addition, we have generated seven constructs using these parts. Furthermore, we have submitted two additional parts representing a foundational tool, including a gene encoding an amber suppressed RFP biobrick for screening of SupF+ (Amber suppressor) genotype and an amber suppressor tRNA biobrick. <br /> |
</p> | </p> | ||
Revision as of 02:01, 30 October 2008
|
|
 "
"