Team:Rice University/CONSTRUCTS
From 2008.igem.org
DavidOuyang (Talk | contribs) |
StevensonT (Talk | contribs) (→Constructs) |
||
| Line 150: | Line 150: | ||
<tr> | <tr> | ||
<td width="71">[http://partsregistry.org/wiki/index.php?title=Part:BBa_K122007 K122007]</td> | <td width="71">[http://partsregistry.org/wiki/index.php?title=Part:BBa_K122007 K122007]</td> | ||
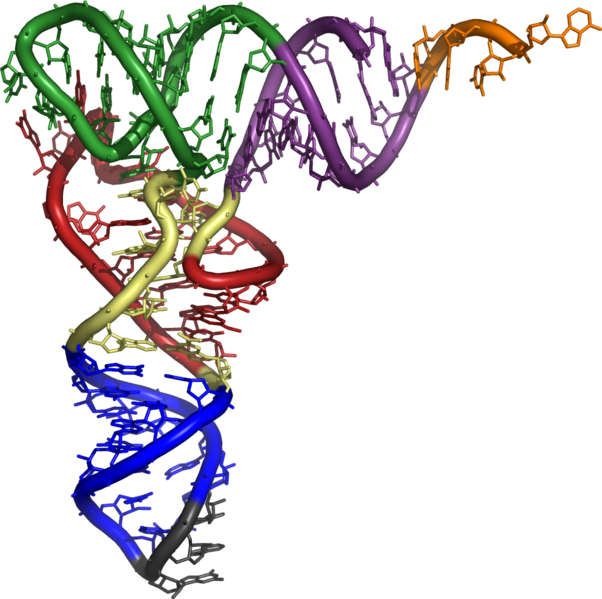
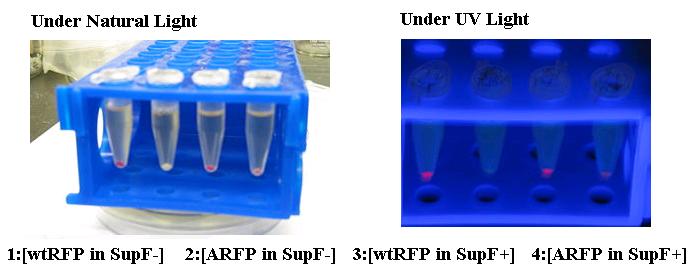
| - | <td width="354">The supF construct, an amber suppressor tRNA, allows for read-through at native amber (TAG) stop codons.</td> | + | <td width="354">The supF construct, an amber suppressor tRNA, allows for read-through at native amber (TAG) stop codons. Charges with tyrosine.</td> |
<td width="30">211</td> | <td width="30">211</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td height="54">[http://partsregistry.org/wiki/index.php?title=Part:BBa_K122006 K122006]</td> | <td height="54">[http://partsregistry.org/wiki/index.php?title=Part:BBa_K122006 K122006]</td> | ||
| - | <td>Point Mutation of RFP(13521) with incorporation of an amber stop codon at the fluorophore.</td> | + | <td>Point Mutation of RFP(13521) with incorporation of an amber stop codon at the native tyrosine required for fluorophore maturation.</td> |
<td>923</td> | <td>923</td> | ||
</tr> | </tr> | ||
Revision as of 03:39, 30 October 2008
|
OUR TEAM ::: SUMMARY ::: INTRODUCTION ::: STRATEGY ::: RESULTS ::: ONGOING WORK ::: GALLERY |
 "
"