Team:Rice University/RESULTS
From 2008.igem.org
(→C. HPLC Data) |
(→HPLC: Calibration) |
||
| Line 49: | Line 49: | ||
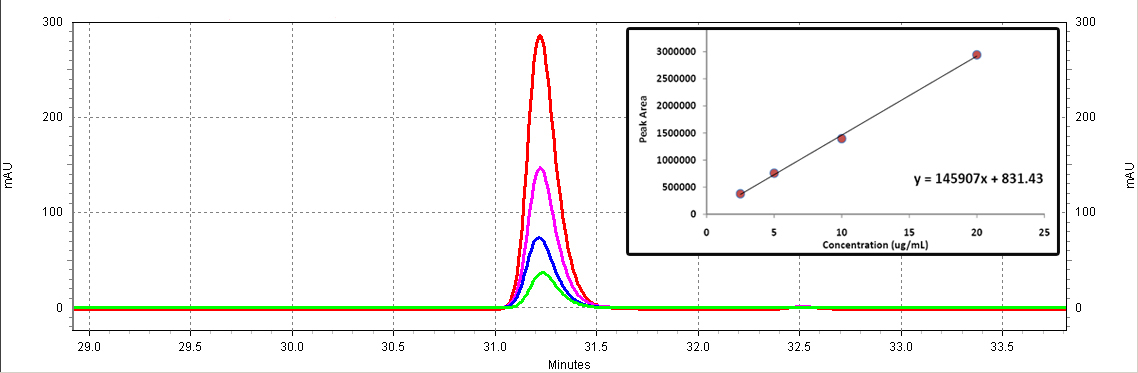
[[Image:Coumaric_standards.jpg|center|thumb|600px|Figure C3: p-Coumaric acid standards (Sigma). Different serial dilutions are shown: 18.5ug/mL (red), 9.25ug/mL (purple), 4.63ug/mL (blue), 2.3ug/mL (green).]]<BR> | [[Image:Coumaric_standards.jpg|center|thumb|600px|Figure C3: p-Coumaric acid standards (Sigma). Different serial dilutions are shown: 18.5ug/mL (red), 9.25ug/mL (purple), 4.63ug/mL (blue), 2.3ug/mL (green).]]<BR> | ||
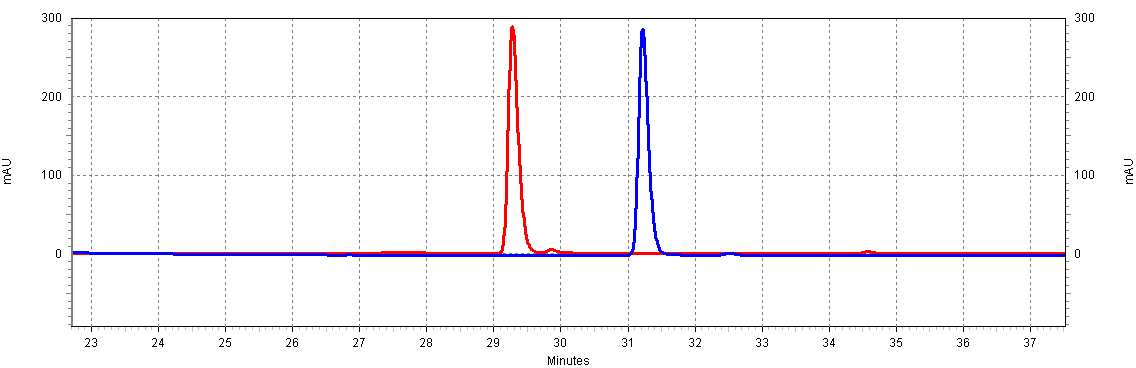
| - | [[Image:RSVandCoumaric.jpg|center|thumb|600px|Figure C4: trans-resveratrol and p-coumaric acid separate at different times; this will allow us to resolve both molecules in a complex mixture, such as a yeast extract.]]<BR>]] | + | [[Image:RSVandCoumaric.jpg|center|thumb|600px|Figure C4: trans-resveratrol and p-coumaric acid separate at different times; this will allow us to resolve both molecules in a complex mixture, such as a yeast extract. Shown is 18.5ug/mL of p-coumaric acid and 20ug/mL trans-resveratrol of ]]<BR>]] |
===== HPLC: Wine Extract Tests===== | ===== HPLC: Wine Extract Tests===== | ||
Revision as of 02:27, 30 October 2008
|
|
 "
"