Team:Tsinghua/Project
From 2008.igem.org
| HOME | Team | Project 1 | Project 2 | Parts | Modelling | Notebook | Doodle Board |
|---|
Contents |
Background
| Motile bacteria can respond to chemical gradients in their environment. Such directed movement in response to special chemicals is defined as chemotaxis. If organisms detect substances beneficial for surviving, positive chemotaxis occurs and cells will move in the direction of increasing concentration. In contrast, negative chemotaxis occurs when motile bacteria move away from a repellant. Both kinds of responses adjust the behaviors of organisms to the environment, thus improve their ability of surviving.
One typical chemotactic behavior is the response of Escherichia coli toward certain nutritions, which plays a key role in seeking food. Researchers have investigated thoroughly into the molecular mechanism of it.Motility in prokaryotes is accomplished by flagellum, a complex structure moving the cell by rotation, much like the propeller in a motor boat. In E.coli, counterclockwise rotation moves the cell in a direction called a run, while clockwise rotation results in tumbling of the cell.The direction of rotation is controlled by phosphorylation of CheY, a signaling protein in the chemotaxis pathway of E.coli. Low level of phosphorylation results in counterclockwise rotation of flagella and makes the cell move forward. In contrast, phosphorylated CheY favors tumbling. The level of protein phosphorylation is regulated by certain attractant or repellent, which can bind specific receptors on the cell surface. Signals are then passed through a series of intracellular biochemical reactions, in which phosphorylation of CheY is controlled. In this way bacteria accomplish chemotactic behavior. Escherichia coli is a common kind of engineer bacteria, which is convenient for genetic manipulation. It is likely that we make use of the chemotactic pathway in E.coli, making them detect and move toward a special chemical substance, maybe a certain kind of pollutant. If the genetic engineering strain also has the ability of degrading aimed chemicals, efficiency of pollution treatment can be hopefully improved a lot. However, the natural ligands concerning chemotaxis in E.coli are limited in several kinds of nutrition, such as carbohydrate and amino acids. Strains that can detecting more kinds of organic substances and even heavy metal ions is planed to be got through the reconstruction of chemotaxis system. |
Overall project
This year we are trying to work at the motility of bacteria. E.coli senses some chemicals and moves toward or stays still, this behavior is called chemotaxis. Any modification of the pathway from the sensor to the motile apparatus will cause different chemotactic effcts.
Moreover,most of the biosensors today are designed for a specific target signal, and they can not substitute each other in most cases. By structural analysis we find that most receptors for the same type of ligands exhibit great similarity, both in sequence information and characteristics. Our project is designed for an "Interchangeable Biosensor". The strategy is almost trial and error, to find an appropriate pathway between chemical molecules and corresponding reactions. Signals other than chemicals are much harder to manipulate, and integrating all the signals seems even more challenging,we are trying to isolate the elements and standardize them.
Major inspiration of our idea comes from the Three Laws of Robotics(Isaac Asimov):
1. A robot may not injure a human being, or, through inaction, allow a human being to come to harm.
2. A robot must obey the orders given it by human beings except where such orders would conflict with the First Law.
3. A robot must protect its own existence as long as such protection does not conflict with the First or Second Law.
The goal is to be achieved via three different subprojects: CdLocalizer, DualReceptor and TheoRiboSwitch.
TheoRiboSwitch Project focuses on the verification of signals transmission between GCS and motor while DualReceptor Project focuses on the integration of multiple signals at the sensor and GCS sections. CdLocalizer Project is designed to illustrate a more complicated correlation other than linear correlation between ligand concentration and GCS.
Standardized Chemokactic Access Control and Modular Design of E.coli
One canonical chemotactic behavior is the response of Escherichia coli toward certain nutrition, which plays a key role in seeking food. Researchers have investigated thoroughly into the molecular mechanism of it.
Motility in prokaryotes is accomplished by flagellum, a complex structure moving the cell by rotation, much like the propeller in a motor boat. In E.coli, counterclockwise rotation moves the cell in a direction called a run, while clockwise rotation results in tumbling of the cell.
The direction of rotation is controlled by phosphorylation of CheY, a signaling protein in the chemotaxis pathway of E.coli. Low level of phosphorylation results in counterclockwise rotation of flagella and makes the cell move forward. In contrast, phosphorylated CheY favors tumbling.
The level of protein phosphorylation is regulated by certain attractant or repellent, which can bind specific receptors on the cell surface. Signals are then passed through a series of intracellular biochemical reactions, in which phosphorylation of CheY is controlled. In this way bacteria accomplish chemotactic behavior.
Molecular mechanism involve in chemotaxis behavior can be simplified into following schematics. There are several kinds of receptors for different chemotactic ligands on the membrane of E.coli. These receptors sense correspondent ligands and convert the concentration of each ligand to their activity. The total activity of them represents whether the bacterium like or hate a certain position in the environment. We name the sum of chemotactic receptors’ activity “General Chemotaxis Signal (GCS)”. Then the decision of chemotaxis is converted into CheY phosphorylation level, which controls rotation bias of molecular motor. Thus, concentration of CheY-p is called “Motor Control Signal (MCS)” here.
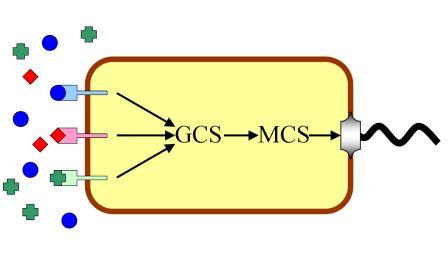
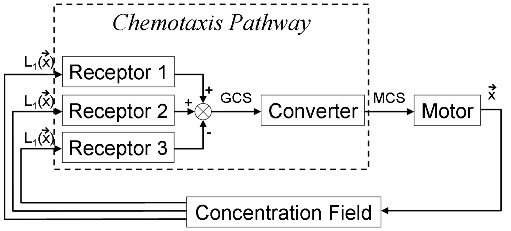 System Block Diagram of Chemotaxis
System Block Diagram of Chemotaxis
Different types of receptors sense the concentrations of various ligands and conform them to General Chemotaxis Signal (GCS), which represents the chemotaxis decision. The rest of chemotaxis pathway is in charge of convert GCS to MCS (Motor Control Signal), which interferences motor rotation bias. The working state of motor are directly related to the positions of bacteria in medium, and coordinate of bacterium corresponds to the concentration of ligands there. Bacteria sense real-time environmental conditions and regulate its movement model through this mechanism in order to find a place with highest fitness.
Status of the Three Experimental Programs in the Entire Study
TheoRiboSwitch Project
This program is mainly used to verify the design principle of signal pathway between GCS and MCS. As we have mentioned, the relation between GCS and MCS is not a simple positive correlation, but one similar to derivative relation. Their signaling pathway not only transfers the signal from some intermediates to another molecule which can interact with the motor, but also completes a similar calculation of derivation. In our study, by means of reverse engineering, we analyze the dynamic characteristics of E. coli natural chemokine pathways, and abstract the key factors which enable E.coli to guide chemokaxis. Then, on the basis of these dynamics, we modify our design for a new pathway, and verify it via computer simulation. As for the design of the final test, it must be directly linked with real E. coli, which is the final objective of this program. In the program, the concentration ligand and Ribo-switch complex corresponds to common chemokactic signals. This design does not involve the signal integration of a variety of substances, and their combination is a fast chemical balance, which minimizes the delay and makes this program a suitable verification for the second half of the entire design.
DualReceptor Project
The main purpose of this program is to integrate the two signals. Although in practice, the two receptors correspond to the same ligand, yet the relationship between the concentration of the ligand they mediate and GCS is just the opposite. Thus, this program can be seen as the integration (or trade-off) of the two signals. The same kind of ligand molecules are used to facilitate quantitative detection, and also to achieve a certain degree of practical significance (see description below). This program is Theophylline chemokaxis-based. A key issue needs to be addressed during the convergence of integration phase and transference phase of the signal: would the delay of signal in the first part of integration impact later signal transference? This requires proof based on theoretical analysis and simulation experiments in advance. Of course, we can also choose to establish only positive correlation between GCS and MCS, which can be called “pseudochemokaxis”, to avoid interference by the progress of the Theophylline Chemokaxis Program.
CdLocalizer Project
In this program, we attempt to establish a more complex function, rather than a simple positive correlation between ligand concentration and GCS. Although the ultimate application purpose of the program is the same with the Toluene Chemokaxis Program, a different design principle is used. Just like the Toluene Chemokaxis Program, this one is faced with the same signal-conversion delay problem. Still, we can use “pseudochemokaxis” to complete signal conversion of the second half of the whole process.
Wetlab Work
Simulation
Reference
1 Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Vivian Hsiu-Chuan Liao*, Ming-Te Chien, Yuen-Yi Tseng, Kun-Lin Ou. Environmental Pollution 142 (2006) 17e23
This literature helped us to introduce GFP into our project and gave us a description of heavy metal pollutants.
2 The C-terminal domain of the Escherichia coli RNA polymerase a subunit plays a role in the CI-dependent activation of the bacteriophage lambda pM promoter. Barbara Kedzierska , Anna Szambowska , Anna Herman-Antosiewicz , David J. Lee , Stephen J.W. Busby2, Grzegorz Wegrzyn1 and Mark S. Thomas3,* Nucleic Acids Research, 2007, Vol. 35, No. 7
This literature gives out a mechanism of CI-Pm promotor interaction.
3 Guiding Bacteria with Small Molecules and RNA.Shana Topp and Justin P. Gallivan. J. AM. CHEM. SOC. 2007, 129, 6807-6811 9
This literature lent us an overall vision of how bacteria movement happens and indicated out the central role of CheZ. It also introduced riboswitch into our project.
4 Luminescent bacterial sensor for cadmium and lead.Sisko Tauriainen, Matti Karp, Wei Chang, Marko Virta.Biosensors & Bioelectronics 13 (1998) 931–938
This literature descripted sensors for cadmium and lead.
5 Online and in situ monitoring of environmental pollutants: electrochemical biosensing of cadmium.I Biran, R Babai, K Levcov, J Rishpon, EZ Ron.Environmental Microbiology, 2000
This literature gives out a sensing and detecting method for cadmium.
6 Sensitivity of OR in Phage lambda.Audun Bakk, Ralf Metzler, and Kim Sneppen.Biophysical Journal Volume 86 January 2004 58–66
This literature explains the operation of operons in phage lambda.
7 An Artificial Aptazyme-Based Riboswitch and its Cascading System in E. coli. Atsushi Ogawa and Mizuo Maeda. ChemBioChem 2008, 9, 206 – 209
This is an indication of riboswitch.
Other reference
Goldstein RA & Soyer OS (2008) PLoS Comput Biol 4, e1000084.
Emonet T & Cluzel P (2008) Proc Natl Acad Sci U S A 105, 3304-3309.
Emonet T, Macal CM, North MJ, Wickersham CE, & Cluzel P (2005) Bioinformatics 21, 2714-2721.
Korobkova E, Emonet T, Vilar JM, Shimizu TS, & Cluzel P (2004) Nature 428, 574-578.
Sourjik V & Berg HC (2002) Proc Natl Acad Sci U S A 99, 123-127.
Yi TM, Huang Y, Simon MI, & Doyle J (2000) Proc Natl Acad Sci U S A 97, 4649-4653.
Lauffenburger DA (2000) Proc Natl Acad Sci U S A 97, 5031-5033.
Liu Z & Papadopoulos KD (1996) Biotechnol Bioeng 51, 120-125.
Ishihara A, Segall JE, Block SM, & Berg HC (1983) J Bacteriol 155, 228-237.
Cluzel P, Surette M, & Leibler S (2000) Science 287, 1652-1655.
| HOME | Team | Project | Parts | Modelling | Notebook | Doodle Board |
|---|
 "
"