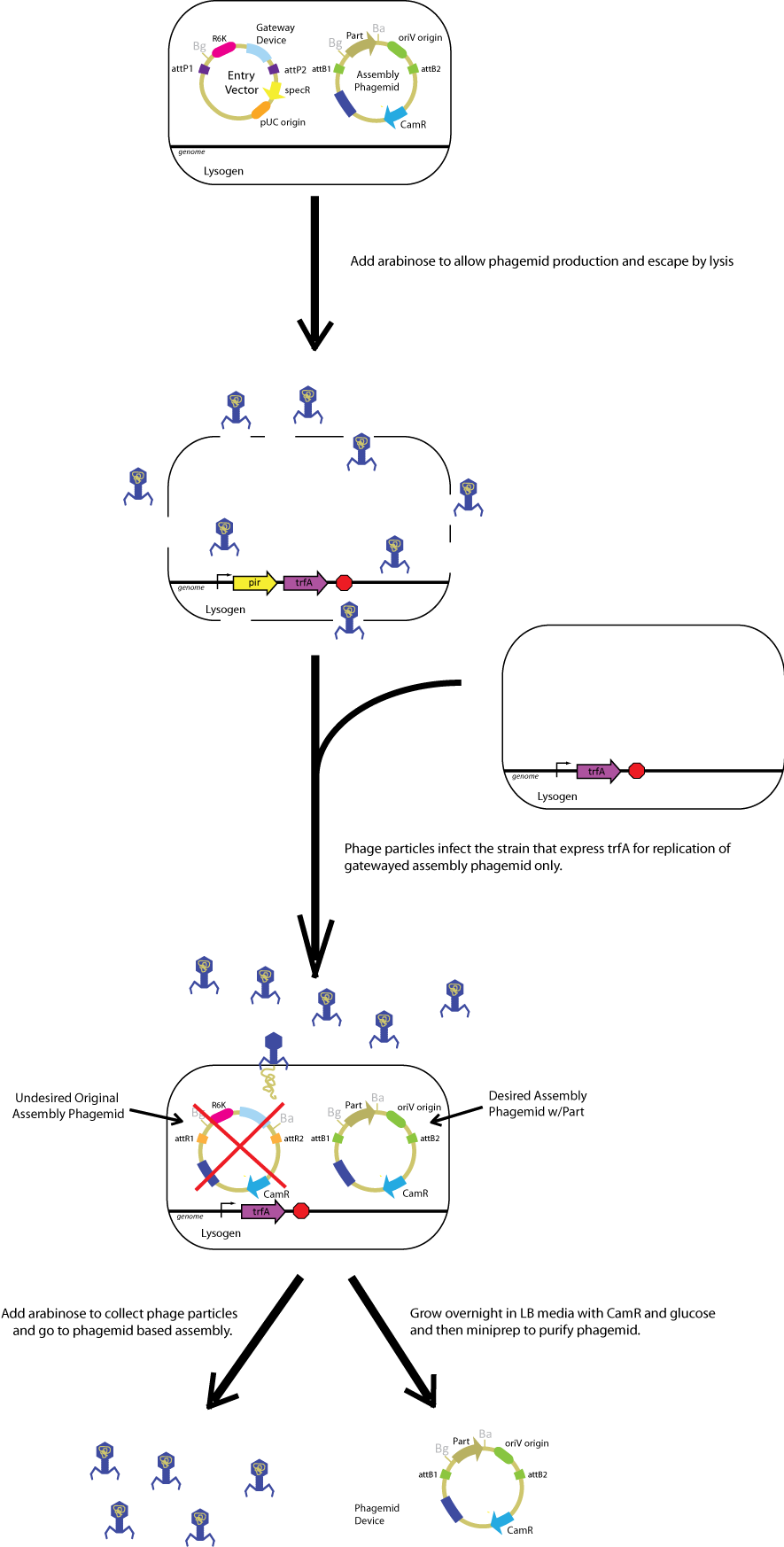Team:UC Berkeley/GatewayPhagemid
From 2008.igem.org
(New page: {{UCBmain|cssLink=}} <div style="text-align: center;"><font size="6">'''Phagemid Based Gateway'''</font></div><br>) |
(→Strains) |
||
| (40 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
{{UCBmain|cssLink=}} | {{UCBmain|cssLink=}} | ||
<div style="text-align: center;"><font size="6">'''Phagemid Based Gateway'''</font></div><br> | <div style="text-align: center;"><font size="6">'''Phagemid Based Gateway'''</font></div><br> | ||
| + | |||
| + | ==Introduction== | ||
| + | The phagemid scheme was motivated by a desire to further streamline the protocol for performing Gateway ''in vivo''. Like the genomic scheme, this method utilizes positive selection to isolate the desired product. However, it improves on this scheme because the lysis and infection steps replace mini-preps and transformations without requiring additional devices. | ||
| + | |||
| + | This scheme utilizes two methods of positive selection: conditional origins of replication (''oriV'' and ''oriR6K'') and the ability to be packaged into and transported by a phage. When induced with arabinose, the phagemid (plasmid that can be packaged and transported by a phage) will be copied and packaged in phages that will subsequently lyse the cell. The resulting lysate containing phages holding the plasmid of interest can be used to transport the plasmid into other cells via infection. | ||
| + | |||
| + | ==Details about Plasmids and Strains== | ||
| + | ===Plasmids=== | ||
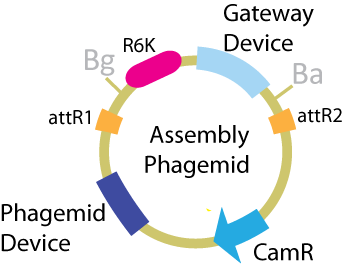
| + | The critical plasmid for this scheme is the phagemid, which contains the assembly vector components as well as the device allowing the plasmid to be packaged and transported by a phage. The components of this plasmid that are transferred to the desired product are the phagemid device and the antibiotic resistance. | ||
| + | |||
| + | [[Image:.ucbphagemid.png|frame|center|Composition of the phagemid.]] | ||
| + | |||
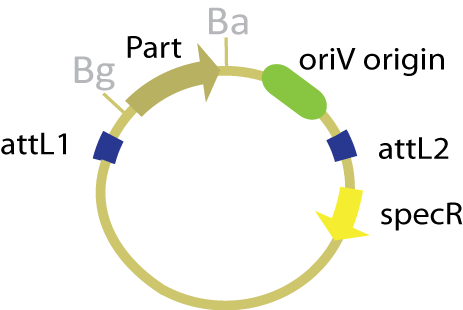
| + | The other plasmid used in this scheme is the ''oriV'' version of the entry plasmid that was used in the genomic scheme. The vector contains the conditional origin ''oriV'' (which is active in the presence of TrfA) as well as the part of interest within the ''attL'' sites. Both the conditional origin and the part are transferred to the assembly vector in the course of the reaction. | ||
| + | |||
| + | [[Image:OriV entry vector.jpg|200 px|frame|center|''oriV'' version of the entry plasmid.]] | ||
| + | |||
| + | ===Strains=== | ||
| + | This method utilizes cells that express TrfA which can activate the ''oriV'' replication origin in the entry plasmid and desired plasmid. It also uses strains that express Pi in order to allow the phagemid to replicate when it is in its plasmid form. Additionally, all of the strains used in the scheme need to be a lysogens (have phage genomic information integrated into their genomes) so that they can create phage particles when they lyse. | ||
| + | |||
| + | ==General Procedure== | ||
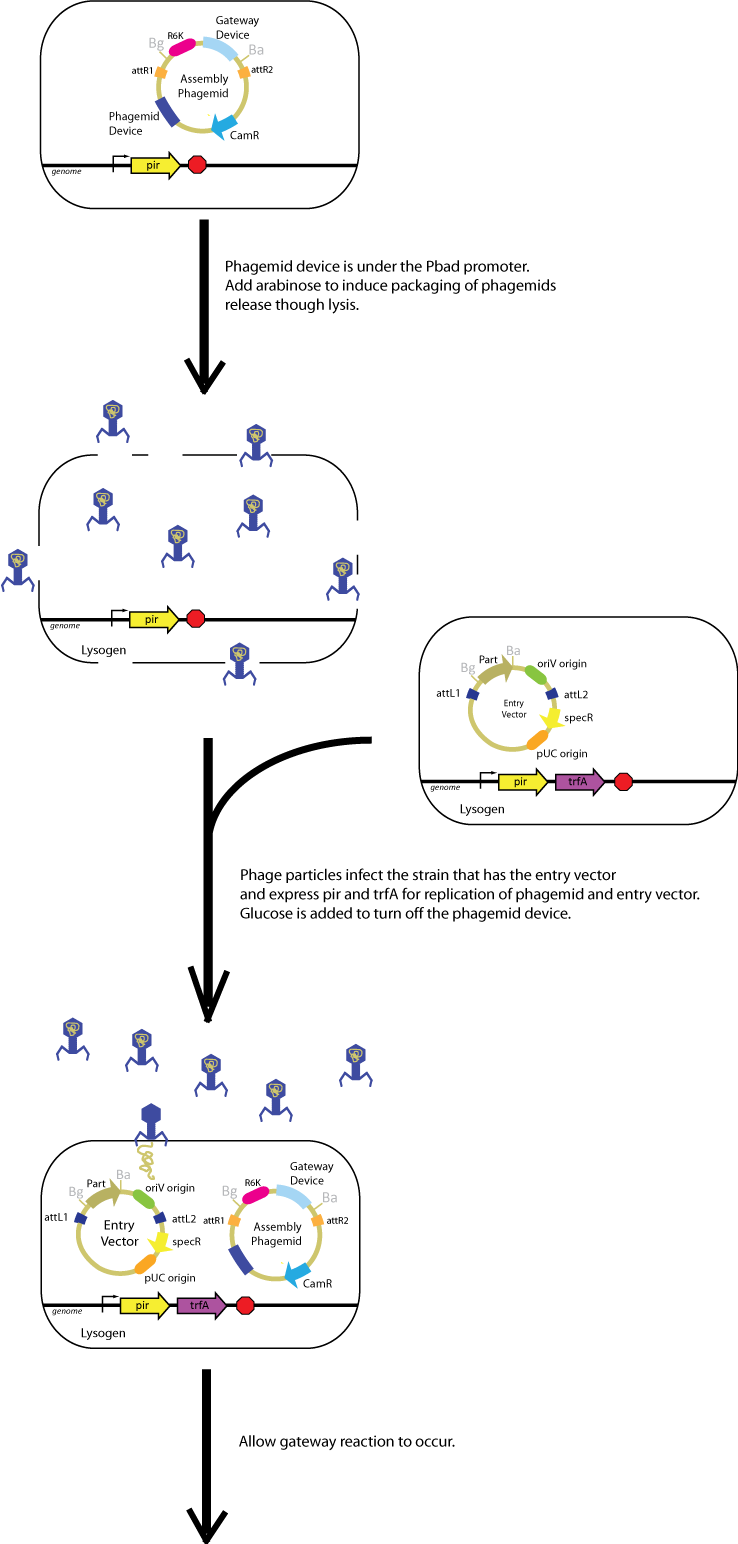
| + | In practice, you will build a Biobrick part in an ''oriV'' entry vector and transform into the strain with ''pir'' and ''trfA'' genes. You can either a) infect these bacteria with phage lysate which contains assembly phagemids packaged in phage or b) introduce purified phagemid DNA directly into the strain with ''oriV'' entry vector by transformation. Here is the brief flow chart of doing phagemid-based Gateway. | ||
| + | |||
| + | [[Image:ucbphagemidoverview1.png|800px]] | ||
| + | [[Image:ucbphagemidoverview2.png|800px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''If you want to see how far we've gotten, come and see our talk!!''' | ||
| + | |||
| + | <html> | ||
| + | <a href="https://2008.igem.org/Team:UC_Berkeley/LysisDevice" class="titleIcon"> | ||
| + | <img align=right src="https://static.igem.org/mediawiki/2008/9/9a/GreyNext.png"> | ||
| + | </a> | ||
| + | </html> | ||
Latest revision as of 04:15, 30 October 2008
Introduction
The phagemid scheme was motivated by a desire to further streamline the protocol for performing Gateway in vivo. Like the genomic scheme, this method utilizes positive selection to isolate the desired product. However, it improves on this scheme because the lysis and infection steps replace mini-preps and transformations without requiring additional devices.
This scheme utilizes two methods of positive selection: conditional origins of replication (oriV and oriR6K) and the ability to be packaged into and transported by a phage. When induced with arabinose, the phagemid (plasmid that can be packaged and transported by a phage) will be copied and packaged in phages that will subsequently lyse the cell. The resulting lysate containing phages holding the plasmid of interest can be used to transport the plasmid into other cells via infection.
Details about Plasmids and Strains
Plasmids
The critical plasmid for this scheme is the phagemid, which contains the assembly vector components as well as the device allowing the plasmid to be packaged and transported by a phage. The components of this plasmid that are transferred to the desired product are the phagemid device and the antibiotic resistance.
The other plasmid used in this scheme is the oriV version of the entry plasmid that was used in the genomic scheme. The vector contains the conditional origin oriV (which is active in the presence of TrfA) as well as the part of interest within the attL sites. Both the conditional origin and the part are transferred to the assembly vector in the course of the reaction.
Strains
This method utilizes cells that express TrfA which can activate the oriV replication origin in the entry plasmid and desired plasmid. It also uses strains that express Pi in order to allow the phagemid to replicate when it is in its plasmid form. Additionally, all of the strains used in the scheme need to be a lysogens (have phage genomic information integrated into their genomes) so that they can create phage particles when they lyse.
General Procedure
In practice, you will build a Biobrick part in an oriV entry vector and transform into the strain with pir and trfA genes. You can either a) infect these bacteria with phage lysate which contains assembly phagemids packaged in phage or b) introduce purified phagemid DNA directly into the strain with oriV entry vector by transformation. Here is the brief flow chart of doing phagemid-based Gateway.
If you want to see how far we've gotten, come and see our talk!!
 "
"