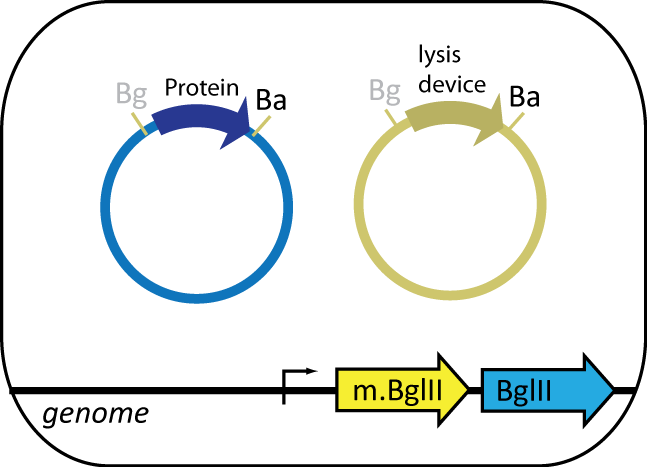Team:UC Berkeley/ProteinPurification
From 2008.igem.org
| Line 31: | Line 31: | ||
'''Tags''' | '''Tags''' | ||
Four tags were made: AP, HA, myc and FLAG. AP binds to streptavidin, Ha binds to hemagglutinin. | Four tags were made: AP, HA, myc and FLAG. AP binds to streptavidin, Ha binds to hemagglutinin. | ||
| + | |||
| + | [[Image:ucbigemtag.png]] | ||
=='''Strategy'''== | =='''Strategy'''== | ||
Revision as of 11:02, 27 October 2008
Contents |
Introduction
Protein purification involves a series of step to isolate the protein of interest. Purifying a protein from E.coli first includes an extraction step, which brings the protein into solution. There are many physical or chemical methods from which to select for extraction. However, many times the potein of interest may be too fragile such that putting the protein through harsh extractions will permanently damage the protein, resulting in low yield. The proposed solution for this is to transform an inducible λ phage lysis device. This gives one complete control over lysis rate. In addition, using a natural form of lysis is much gentler on the cells and proteins, which will lead to greater success of protein extraction and purifcation.
Additional steps can be taken to lower the background that comes from genomic DNA and RNA that flow into solution from extraction. Engineering the ability to make the restriction enzymes BamHI and BgIII and ribonuclease barnase will greatly reduce background.
To identify the protein in the large soup of protein, the four protein tags AP, HA, myc and flag were made. These tags can be attached at the terminus of the protein strands and their selective interactions between antibodies and themselves will make the tagged proteins easier to isolate.
Devices
Expression Cell
The expression cell, containing the plasmid coding for the protein of interest, features BgIII restriction enzyme and BgIII methyltransferase parts engineered into the genome. BgIII, integrated at the HK022 site, is used to cut up the genome of the helper cell. BgIII methyltransferase, integrated at φ80 site, prevents BgIII from cutting up the cell's own genome. The expression cell also contains an inducible lysis device that lyses the cell upon induction.
Helper Cell
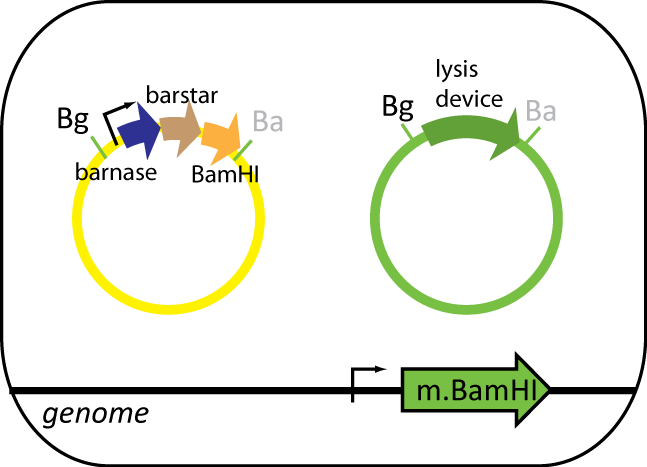
The helper cell contains parts the serve to reduce background during protein purification. The cell has BamHI methyltransferse engineered into its genome at the φ80 site. The plasmid in this cell contains BamHI, barnase and barstar. BamHI is used to degrade the genomic DNA from the expression cell. Barnase is used to degrade RNA of the expression cell and other helper cells when lysed. Barstar prevents barnase from destroy the cell's RNA. Like the expression cell, the helper cells contains the same inducible lysis device.
Protein of Interest
The protein of interest is on a plasmid that is transformed into the expression cell. The protein will have a protein tag.
Tags Four tags were made: AP, HA, myc and FLAG. AP binds to streptavidin, Ha binds to hemagglutinin.
Strategy
The protein purification strategy involves growing up cultures of expression cells and helper cells. Combining the two cultures and lysing them will result in a solution of proteins, among which contains the protein of interest and cellular junk minus the RNA (degraded by barnase) and DNA (degraded by one of the two restriction enzymes). At this point, using the tag on the protein, the protein will be taken out of solution by taking advantage of the selective interaction between the tag and the antibody that binds to it.
Proof of Concept Experiments
To prove the protein purification concept, a plasmid coding for a protein with a His tag is transformed into E.coli with the arabinose induced self-lysis device. This culture will be induced to lyse with arabinose, releasing the His-tagged protein. The cellular debris is pelleted by centrifuging for 30 seconds. The supernatant will be added to a new tube and Ni-NTA will be added to it. The supernatant and Ni-NTA solution is incubated at 4 degree Celcius for 4 hours. The Ni-NTA protein complex is concentrated using a magnet. The protein is eluted from Ni-NTA and the solution is ran on a gel to see whether or not protein is purified.
 "
"