Team:UC Berkeley Tools/Development
From 2008.igem.org
Clotho Beta: Future Development
The next release of Clotho after iGEM is planned for a late Spring 2009 release. This will be the official Beta release (the other releases will have been Alpha and iGEM). The main focus of the release will be to fully implement all the features currently in Clotho as well as fix bugs that are found. Clotho testing and deployment will begin with Anderson Lab at UCB and expand to Voigt Lab at UCSF. In addition to general robustness, Clotho Beta should also incorporate a few new features.
New Features
- New design views to supplement the Sequence View and allow easier designing of basic and composite BioBrick parts. This will likely include a 'schematic view' for assembling parts, similar to the one in [http://web.mit.edu/jagoler/www/biojade/overview.html BioJade].
- Ability to interface with devices, including robots that can be used for automated production of parts designed in Clotho. (In the future, this will contain a link to Anderson Lab's Biomek robot.)
Improved Features:
- More database support: we plan to expand the number and variety of databases that Clotho can connect to, including [http://brickit.wiki.sourceforge.net/ BrickIt!]) and the JBEI registry. One of the databases at the top of our priority list is iGEM's own [http://partsregistry.org/Main_Page Registry of Standard Biological Parts].
- Improved Plug-in environment that allows users to add even more expanded functionality to Clotho. This will include more extension points where plug-ins can be integrated and a set of more fully featured interfaces for existing extensions.
Helping Out In The Lab: Assembly Algorithms
- One of the initial algorithms we added to Clotho was an "optimal assembly" algorithm that can take in a list of goal composite parts and return a list with the minimum number of steps required to build those parts - a particularly useful feature if the goal parts utilize many of the same basic parts as subcomponents.
- EXPAND HERE **
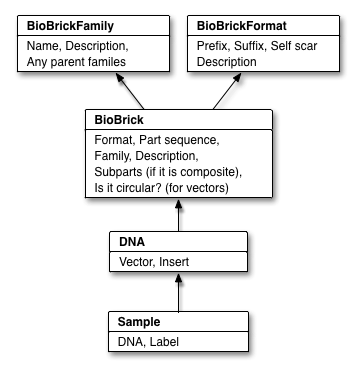
PoBoL: BioBrick Data Sharing Made Easy
PoBoL stands for "Provisional BioBrick Language", and is the name given to a technical standard for BioBrick data in synthetic biology databases. Put simply, our implementation of PoBoL
For more information on the PoBoL standard, check out [http://www.pobol.org/ www.pobol.org]
Increasing Collaboration Through Better Access To Database
Through a combination of PoBoL
 "
"