Team:UC Berkeley Tools/Notebook/Anne's Notebook
From 2008.igem.org
Contents |
Personal Info
Name: Anne Van Devender
Home University: Washington and Lee University
Position: Team member, UC_Berkeley_Tools
Supervisor: Doug Densmore
Lab coworkers: Matthew Johnson, Nade Sritanyaratana
Log
Week One--June 9th
This week was my first working on Berkeley's iGem Computational Team. The week was spent mostly getting background information about the topic. I knew very little about biology and, much less, synthetic biology coming into this project, but with the help of Nade and Matt as well as Professor Anderson, I am beginning to understand the process. With my introduction to biology, I was also introduced to various computational tools used in the field of synthetic biology like BioJade, APE, and GenoCAD. Our tool, Clotho, will have many of the features of these tools in addition to others.
Specifically, the project that I will be working on will be to add an assembly feature to our software. This feature will be used to input a specific set of goal parts and return an output of all possible subparts both in text and graphical display. This result will be achieved through the implementation of an algorithm, which optimally assembles a goal part.
Next week, I will begin implementing this algorithm as well as constructing the basic design of my "Assembly View".
Week Two--June 16th
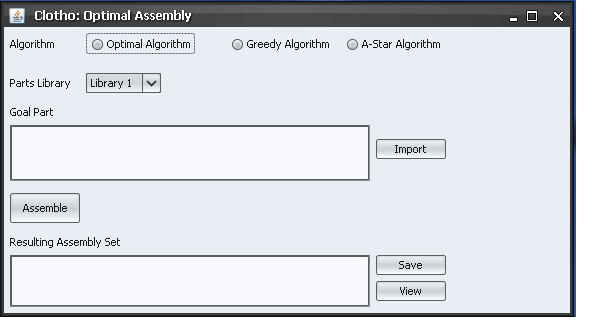
I began this week my creating the view I wanted for assembling parts. Below I have included a screen shot of the assembly window as well as a description of each feature.
- --The Algorithm radio button choices are not actual, but merely examples of the different options you might choose to use for assembly.
- --The Parts library drop-down would allow the user to choose a library from which parts could be accessed, for example, the MIT Parts Repository.
- --The "Goal Part" text area would allow the user to either type directly into the text area the goal part(s) they want to use or they could use the "Import" button to import the goal part from another source (an Excel document, for example).
- --The "Assemble" button would actually perform the algorithm on your specified goal part and place that result in the "Resulting Assembly Set" text area.
- --The "Resulting Assembly Set" text area would have a text-based representation of the assembly sets resulting from the goal parts.
- -- The "Save" button would save this text in a text file to be referenced later.
- --The "View" button would show a graphical representation of the assembly tree(s).
Currently, I am working on the implementation of the algorithm as well as corresponding with a fellow undergraduate, Will DeLoache, about the modification of the algorithm to
include antibiotics. Because of my lack of knowledge of biology, the inclusion of antibiotics was a problem I had not anticipated. Basically, every subpart
would have a specific antibiotic associated with it, and that antibiotic would determine what the resulting composite part's antibiotic would be. This introduction changes the optimization of the algorithm as a common subpart now has 6 different varieties (the 6 different combinations
of antibiotics) associated with each. To find the most optimal subpart for reuse the algorithm now has to take into account the antibiotic varieties of each common subpart. For example, if part ab is used 5 times in assembling a goal part, with three of those being ab/Antibiotic1 and Antibiotic2
the other 2 being ab/Antibiotic2 and Antibiotic3, then ab/Antibiotic1 and Antibiotic2 would be the optimal choice for reuse.
Next week, I will hopefully finish the implementation of both algorithms, and begin working on different forms of input and output for goal parts and resulting assembly sets.
Week Four: June 30th
This week the tool transformed from “Optimal Assembly” to “Algorithm Manager” as we realized it would be more useful if it were more flexible. The tool still works with the two assembly algorithms we have written (WOAA and Optimal Assembly), but it can be extended to include algorithms that have nothing to do with assembly. Extending it in this way, also allows for easier integration into Clotho. Basically, with a few lines of code anyone can add their own algorithm.
This flexibility is achieved through the implementation of an abstract Algorithm class. This class currently contains three different abstract methods which all subclasses of “Algorithm” must implement: getInput(String), getOutputToString(), and run(). Basically, in order for an algorithm to be workable in Clotho it must be able to take a String input that is retrieved from the text area in the GUI, process that data with run(), and return the result in the form of a String to be displayed in the output text area. In reality, the algorithm in not required to take any input or return any output, but for interfacing with the GUI these are the requirements.
We are hoping this flexibility will allow us to implement different types of algorithms in the future.
Week Four: July 7th
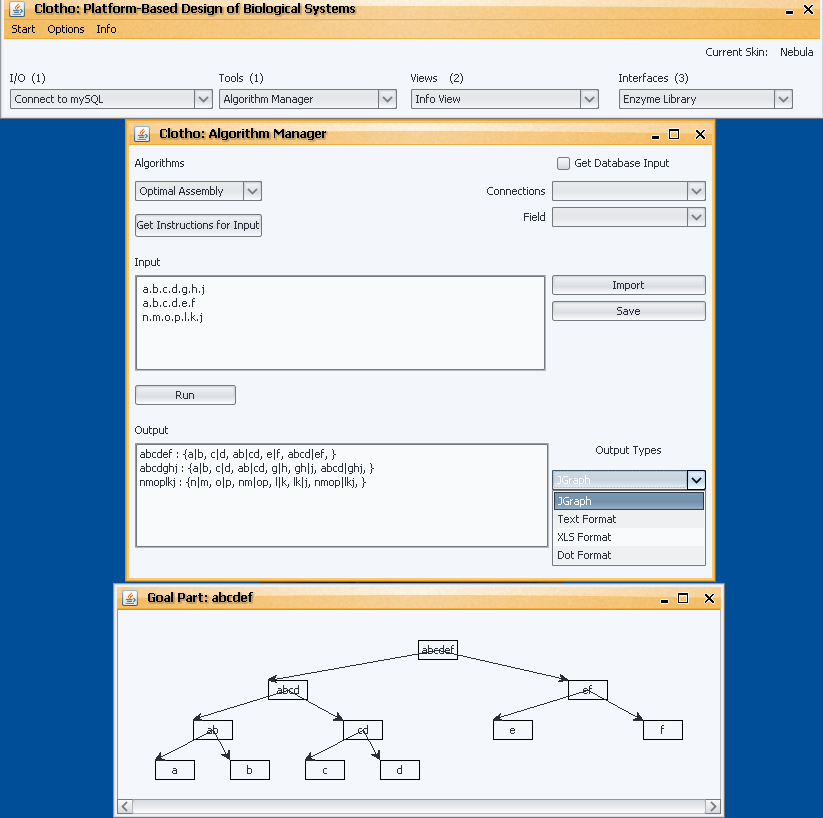
After generalizing my tool to be an Algorithm Manager, I have also made it dynamic in the sense that now instead of selecting algorithms from different radio buttons, a JComboBox is populated with whatever algorithms are integrated in the tool at that time. I have also added another dynamic aspect in that the possible outputs for each algorithm are also dynamically populated upon choosing that algorithm. This feature is achieved by adding an additional abstract method to the Algorithm class that requires each algorithm to tell what types of outputs it supports. For example, the Optimal Algorithm supports JGraph, Excel, Dot, and Text, while WOAA supports Excel and Dot. When you select “Optimal Algorithm” from the algorithms combo box the outputs combo box is dynamically populated with the corresponding outputs.
The last dynamic feature I have added is a set of instructions associated with each algorithm. Again, an abstract method called “getInstructions()” was added to the Algorithm class requiring each subclass to set a set of instructions for its algorithm. This feature was designed to give the user information about the specified algorithm and what type of format you should enter your input in. Depending upon the algorithm you have chosen a different set of instructions will pop up when you select the “Get Instructions for Input” button.
I am happy with the new product, and think it is an improvement over our more limiting, original tool. The basics of the tool seem to be done at this point, although I have a few features in mind that I would like to implement, namely a connection to databases.
Week Four: July 14th
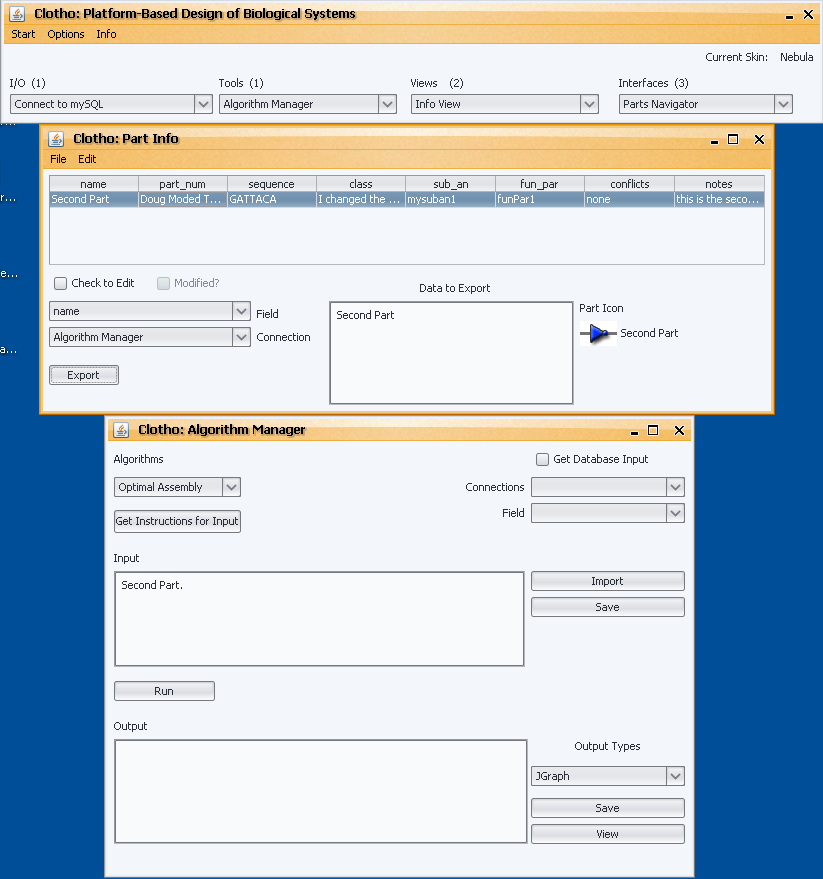
This week was a clean-up. Monday and Wednesday were our testing days. The Algorithm Manager did not get as much feedback and the Sequence View, however, so I was left with fewer bugs. Tuesday was spent trying to add the database connections that I talked about in my last post. Working with Clotho Connections and the ClotoCore, it was fairly easy to exchanged data between the mySQLConnection and the AssemblyConnection.
By Thursday, I had the Algorithm Manager talking to the database. You could export parts from the database straight into the input area of the Algorithm Manager. I also added another method to the Algorithm class called “processDatabaseData(String)” in order to take this sort of action into account. Now, an algorithm can take that individual input and format it in its own specified format. This method was added to ease use as now the user does not have to remember the specific format, she can simply export and everything is done automatically in the Algorithm Manager.
Next week will be spent doing final testing as well as making sure the documentation is clear and complete.
 "
"