Team:UC Berkeley Tools/Notebook/Anne's Notebook
From 2008.igem.org
| Home | The Team | The Project | Notebook |
|---|
Contents |
Personal Info
Name: Anne Van Devender
Home University: Washington and Lee University
Position: Team member, UC_Berkeley_Tools
Supervisor: Doug Densmore
Lab coworkers: Matthew Johnson, Nade Sritanyaratana
Log
Week One--June 9th
This week was my first working on Berkeley's iGem Computational Team. The week was spent mostly getting background information about the topic. I knew very little about biology and, much less, synthetic biology coming into this project, but with the help of Nade and Matt as well as Professor Anderson, I am beginning to understand the process. With my introduction to biology, I was also introduced to various computational tools used in the field of synthetic biology like BioJade, APE, and GenoCAD. Our tool, Clotho, will have many of the features of these tools in addition to others.
Specifically, the project that I will be working on will be to add an assembly feature to our software. This feature will be used to input a specific set of goal parts and return an output of all possible subparts both in text and graphical display. This result will be achieved through the implementation of an algorithm. Currently the algorithm we will be using was
Chris describes four diffrent approaches to building goal parts in his paper: brute-force, top-down, bottom-up, and common-subpart use. The first three are fairly self explanatory, while the last involves constructing a goal part while keeping track of the use of common-subparts. A common-subpart being described as any subpart that is used more than once in a goal part or set of goal parts. Although not always optimal, in many cases buildling a subpart that will be used multiple times in the construction of a goal part, can optimize time and save stages and/or steps in construction.
Next week, I will begin implementing this algorithm as well as constructing the basic design of my "Assembly View". I anticipate keeping in close correspondence with Chris in order to understand his algorithm fully and achieve the best possible implementation in Java.
Week Two--June 16th
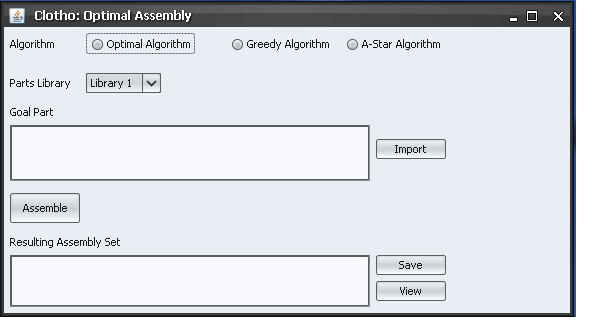
I began this week my creating the view I wanted for assembling parts. Below I have included a screen shot of the assembly window as well as a description of each feature.
- --The Algorithm radio button choices are not actual, but merely examples of the different options you might choose to use for assembly.
- --The Parts library drop-down would allow the user to choose a library from which parts could be accessed, for example, the MIT Parts Repository.
- --The "Goal Part" text area would allow the user to either type directly into the text area the goal part(s) they want to use or they could use the "Import" button to import the goal part from another source (an Excel document, for example).
- --The "Assemble" button would actually perform the algorithm on your specified goal part and place that result in the "Resulting Assembly Set" text area.
- --The "Resulting Assembly Set" text area would have a text-based representation of the assembly sets resulting from the goal parts.
- -- The "Save" button would save this text in a text file to be referenced later.
- --The "View" button would show a graphical representation of the assembly tree(s).
Currently, I am working on the implementation of Chris's algorithm as well as corresponding with a fellow undergraduate, Will DeLoache, about the modification of Chris's algorithm to
include antibiotics. Because of my lack of knowledge of biology, the inclusion of antibiotics was a problem I had not anticipated. Basically, every subpart
would have a specific antibiotic associated with it, and that antibiotic would determine what the resulting composite part's antibiotic would be. This introduction changes the optimization of the algorithm as a common subpart now has 6 different varieties (the 6 different combinations
of antibiotics) associated with each. To find the most optimal subpart for reuse the algorithm now has to take into account the antibiotic varieties of each common subpart. For example, if part ab is used 5 times in assembling a goal part, with three of those being ab/Antibiotic1 and Antibiotic2
the other 2 being ab/Antibiotic2 and Antibiotic3, then ab/Antibiotic1 and Antibiotic2 would be the optimal choice for reuse.
Next week, I will hopefully finish the implementation of both algorithms, and begin working on different forms of input and output for goal parts and resulting assembly sets.
 "
"