Team:UC Berkeley Tools/Project/Tutorial/SequenceView2
From 2008.igem.org
| Line 7: | Line 7: | ||
| - | + | <p><br><br>[[Team:UC_Berkeley_Tools/Project/Tutorial/RestrictionSiteHighlighting|'''<<< Previous''']] | [[Team:UC_Berkeley_Tools/Project/Tutorial/Database|'''Next >>>''']]</p> | |
<p><strong>6) Sequence View II</strong><br /> | <p><strong>6) Sequence View II</strong><br /> | ||
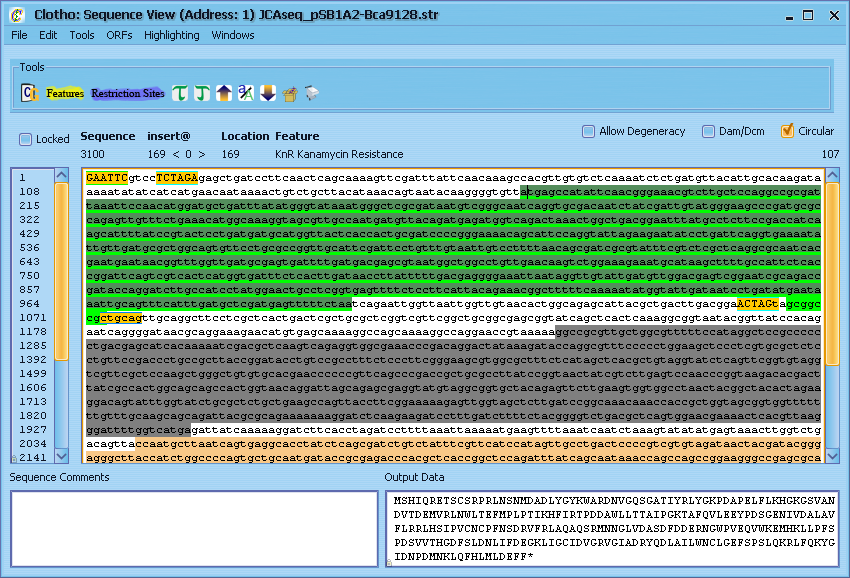
Now let's play with some of the other basic tools in the Sequence View. First, let's find an open reading frame - place your cursor anywhere in the sequence, go to the "ORFs" menu and select "Find Next ORF". Alternatively, you can use the shortcut and press [Ctrl + .]. This will place line a light green line underneath the next open reading frame found in the sequence after the cursor (if other alternate start codons are enabled in the preferences, their reading frames will be shown using yellow or red lines). Highlight this sequence with the mouse and select the "Translate" option from the "Tools" menu (or press [Ctrl + T]). The protein translation of this reading frame will be printed to the 'Output Data' window. Similiarly, if you select "Reverse Complement" [Ctrl + /], the reverse complement of the sequence will be printed. "Find Reverse ORF" and "Reverse Translate" operate in the same fashion, but they look at the reverse complement of the sequence. "UPPER CASE", "Switch Case", and "lower case" all operate on any portion of the sequence you care to highlight (case has no effect on sequence analysis and is treated as aesthetic). Highlighting a string of nucleotides also displays information like G-C content and melting temperature. Additionally, you can open up a tools menu under "Tools" [Ctrl + F] to peform find and replace operations on the sequence. Let's leave the Sequnce View for now - either select "Exit Sequence View" from the File menu or click the 'X' in the corner and select "Yes close" from the resulting pop-up box.</p> | Now let's play with some of the other basic tools in the Sequence View. First, let's find an open reading frame - place your cursor anywhere in the sequence, go to the "ORFs" menu and select "Find Next ORF". Alternatively, you can use the shortcut and press [Ctrl + .]. This will place line a light green line underneath the next open reading frame found in the sequence after the cursor (if other alternate start codons are enabled in the preferences, their reading frames will be shown using yellow or red lines). Highlight this sequence with the mouse and select the "Translate" option from the "Tools" menu (or press [Ctrl + T]). The protein translation of this reading frame will be printed to the 'Output Data' window. Similiarly, if you select "Reverse Complement" [Ctrl + /], the reverse complement of the sequence will be printed. "Find Reverse ORF" and "Reverse Translate" operate in the same fashion, but they look at the reverse complement of the sequence. "UPPER CASE", "Switch Case", and "lower case" all operate on any portion of the sequence you care to highlight (case has no effect on sequence analysis and is treated as aesthetic). Highlighting a string of nucleotides also displays information like G-C content and melting temperature. Additionally, you can open up a tools menu under "Tools" [Ctrl + F] to peform find and replace operations on the sequence. Let's leave the Sequnce View for now - either select "Exit Sequence View" from the File menu or click the 'X' in the corner and select "Yes close" from the resulting pop-up box.</p> | ||
[[Image:Tut6.PNG|center]] <p> </p> | [[Image:Tut6.PNG|center]] <p> </p> | ||
| + | |||
| + | <p>[[Team:UC_Berkeley_Tools/Project/Tutorial/RestrictionSiteHighlighting|'''<<< Previous''']] | [[Team:UC_Berkeley_Tools/Project/Tutorial/Database|'''Next >>>''']]</p> | ||
Latest revision as of 01:44, 24 October 2008
6) Sequence View II
Now let's play with some of the other basic tools in the Sequence View. First, let's find an open reading frame - place your cursor anywhere in the sequence, go to the "ORFs" menu and select "Find Next ORF". Alternatively, you can use the shortcut and press [Ctrl + .]. This will place line a light green line underneath the next open reading frame found in the sequence after the cursor (if other alternate start codons are enabled in the preferences, their reading frames will be shown using yellow or red lines). Highlight this sequence with the mouse and select the "Translate" option from the "Tools" menu (or press [Ctrl + T]). The protein translation of this reading frame will be printed to the 'Output Data' window. Similiarly, if you select "Reverse Complement" [Ctrl + /], the reverse complement of the sequence will be printed. "Find Reverse ORF" and "Reverse Translate" operate in the same fashion, but they look at the reverse complement of the sequence. "UPPER CASE", "Switch Case", and "lower case" all operate on any portion of the sequence you care to highlight (case has no effect on sequence analysis and is treated as aesthetic). Highlighting a string of nucleotides also displays information like G-C content and melting temperature. Additionally, you can open up a tools menu under "Tools" [Ctrl + F] to peform find and replace operations on the sequence. Let's leave the Sequnce View for now - either select "Exit Sequence View" from the File menu or click the 'X' in the corner and select "Yes close" from the resulting pop-up box.
 "
"