Team:University of Lethbridge/Modeling
From 2008.igem.org
(added references) |
(added in BCT link) |
||
| Line 36: | Line 36: | ||
For our project, we decided to create a synthetic bacterium which could move towards and degrade an environmental pollutant. To direct the movement of our modified "Bacuum Cleaner," we reprogrammed the chemotaxis pathway by using a toxin-responsive riboswitch to control expression of the motility protein CheZ. CheZ catalyzes the dephosphorylation of the motility protein CheY, which promotes directed movement (chemotaxis) over the "tumbling" state of the bacterium. | For our project, we decided to create a synthetic bacterium which could move towards and degrade an environmental pollutant. To direct the movement of our modified "Bacuum Cleaner," we reprogrammed the chemotaxis pathway by using a toxin-responsive riboswitch to control expression of the motility protein CheZ. CheZ catalyzes the dephosphorylation of the motility protein CheY, which promotes directed movement (chemotaxis) over the "tumbling" state of the bacterium. | ||
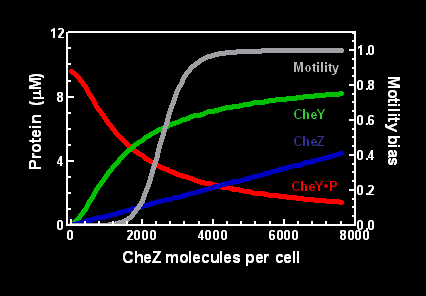
| - | We used the modeling program BCT ( | + | We used the modeling program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html <font color=yellow>BCT</font>] (Bray et al, 1993, 2007; Bray and Bourret, 1995) to determine the level of ''CheZ'' gene expression necessary to produce dephosphorylated CheY and directed movement. Increasing intracellular concentrations of CheZ leads to increased CheY in the unphosphorylated state (Figure 1)... |
---- | ---- | ||
Revision as of 16:53, 29 October 2008
Our Project
For our project, we decided to create a synthetic bacterium which could move towards and degrade an environmental pollutant. To direct the movement of our modified "Bacuum Cleaner," we reprogrammed the chemotaxis pathway by using a toxin-responsive riboswitch to control expression of the motility protein CheZ. CheZ catalyzes the dephosphorylation of the motility protein CheY, which promotes directed movement (chemotaxis) over the "tumbling" state of the bacterium.
We used the modeling program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html BCT] (Bray et al, 1993, 2007; Bray and Bourret, 1995) to determine the level of CheZ gene expression necessary to produce dephosphorylated CheY and directed movement. Increasing intracellular concentrations of CheZ leads to increased CheY in the unphosphorylated state (Figure 1)...
Figure 1
References
Bray, D., Bourret, R. B., and Simon, M. I. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol. Biol. Cell 4, 469-482.
Bray, D., and Bourret, R. B. 1995. Computer analysis of the binding reactions leading to a transmembrane receptor-linked multiprotein complex involved in bacterial chemotaxis. Mol. Biol. Cell 6, 1367-1380.
Bray, D., Levin, M. D., and Lipkow, K. 2007. The chemotactic behavior of computer-based surrogate bacteria. Curr. Biol. 17, 12-19.
</div>
 "
"

 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]