Team:University of Lethbridge/Notebook/GeneralLabAugust
From 2008.igem.org
Munima.alam (Talk | contribs) m (some spelling and spacing - I have issues) |
Munima.alam (Talk | contribs) m |
||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Team:University_of_Lethbridge/Notebook|Back to The University of Lethbridge Main Notebook]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===August 5, 2008=== | ===August 5, 2008=== | ||
| Line 46: | Line 29: | ||
Gel extracted LacI insert and DT vector. Didn't run gel yet. | Gel extracted LacI insert and DT vector. Didn't run gel yet. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 99: | Line 55: | ||
Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes. | Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
====Christa, Munima, Sebastian==== | ====Christa, Munima, Sebastian==== | ||
| Line 160: | Line 66: | ||
Conclusion: The bands appeared to be at the correct size for pSB1A7. | Conclusion: The bands appeared to be at the correct size for pSB1A7. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 194: | Line 85: | ||
| - | ===August | + | ===August 26, 2008=== |
| - | ==== | + | ====Roxanne, John==== |
| - | -Digested | + | -Restriction Digested the Biobrick Parts I13504, I13401, P0440, C0014, B0015, J31007 with XbaI and PstI |
| + | |||
| + | -20 uL template (~2 ug) | ||
| + | -5 uL React 2 | ||
| + | -4 uL XbaI | ||
| + | -4 uL PstI | ||
| + | -17 uL ddH2O | ||
| + | ____ | ||
| + | 50uL Reaction in 37.0C H2O bath overnight | ||
| + | |||
| + | |||
| + | ===August 27, 2008=== | ||
| + | ====Roxanne==== | ||
| + | -Inactivated the Restriction Enzymes by placing them on the heating block at 65.0C for 10 minutes | ||
| + | -Ran 25uL of DNA on a 1% Agarose Gel at 100 V for 30 minutes | ||
| + | -Gel Extracted the DNA | ||
| + | -Ran 1 uL of DNA in a 1% Agarose Gel at 100V for 30 minutes. | ||
| + | |||
| + | [[Image:Biobrick_Parts.jpg| 250 px]] | ||
| + | |||
| - | + | ===August 28, 2008=== | |
| - | + | ====Roxanne, Munima, Sebastian, Nathan Puhl==== | |
| - | + | -Performed a restriction digest on LacI+dt, pLacI, pStrong, and RFP sub | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | - | + | -10 uL template |
| + | -5 uL NEB 2 | ||
| + | -2 uL Restriction Enzyme #1 (Xba I or Spe I) | ||
| + | -2 uL Restriction Enzyme #2 (Pst I) | ||
| + | -21 uL ddH20 | ||
| + | ____ | ||
| + | 50 uL Rxn left to run overnight at 37.0 | ||
| - | -Gel | + | -Ran a Gel of the Riboswitch PCR which had been done with Taq a couple of weeks ago to quantify the amount of DNA present. Setup the Math to perform a ligation into pGEM T-easy. |
| - | - | + | -Picked a colony from the dT plate which was stored in the fridge from several weeks ago, incubated overnight at 37.0C |
| - | |||
| - | - | + | ===August 29, 2008=== |
| + | ====Roxanne, Nathan Puhl, Munima, Sebastian, Andrew==== | ||
| + | -Performed the ligation of the riboswitch into pGEM T-easy, transformed and plated. | ||
| - | + | =====Roxanne==== | |
| - | + | -Gel Extracted the remainder of the DNA | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===August | + | ===August 30, 2008=== |
| - | ====Nathan Puhl==== | + | ====Nathan Puhl, Roxanne, Andrew==== |
| - | - | + | -Ran a gel of the Digested Parts on 1% Agarose @ 100V for 27 minutes. |
Latest revision as of 02:29, 30 October 2008
Back to The University of Lethbridge Main Notebook
Contents |
August 5, 2008
Nathan Puhl et al
Gel extracted two bands really closer to each other from PCR (August 1) at ~76 bp. Used Qiagen MiniElute kit. Results are in the hard copy lab notebook. Seems as if gel extraction was successful. ____ 2 color reporter system:
Set up digest of LacI and DT.
LacI - EcoRI and SpeI
DT - EcoRI and XbaI
Reaction mixture:
-5 uL template -5 uL NEB 2 Buffer -0.5 uL BSA? -1 uL RE #1 -1 uL RE #2 - 37.5 uL ddH20
Left at 37 C overnight. Heat deactivation (65 C) for 15 minutes.
August 6, 2008
Nathan Puhl, Roxanne
2 color reporter system:
Gel extracted LacI insert and DT vector. Didn't run gel yet.
August 13, 2008
Nathan Puhl and Sebastian
Verified and quantified the LacI and DT restriction digest products.
Set up a Ligase experiment for LacI and DT
- 1.3 uL vector(DT) - 8 uL insert(LacI) - 2 uL 10x Ligase buffer - 1.5 uL Ligase - 7.2 uL ddH2O - 20 uL total volume
August 14, 15
Nathan Puhl, Munima, Selina
Poured 61 LB + Amp plates. Stored in iGEM 4 C fridge.
August 15, 2008
Roxanne
Used Qiagen Plasmid MiniPrep Kit to Plasmid Prep Last Year's Biobrick Parts that were Incubated overnight.
Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes.
Christa, Munima, Sebastian
Did a plasmid prep on pSB1A7 using QIAprep Spin Miniprep Kit. Stocked 4- 50uL of pSB1A7 and stored in iGEM -20 C.
Sebastian, Nathan Puhl
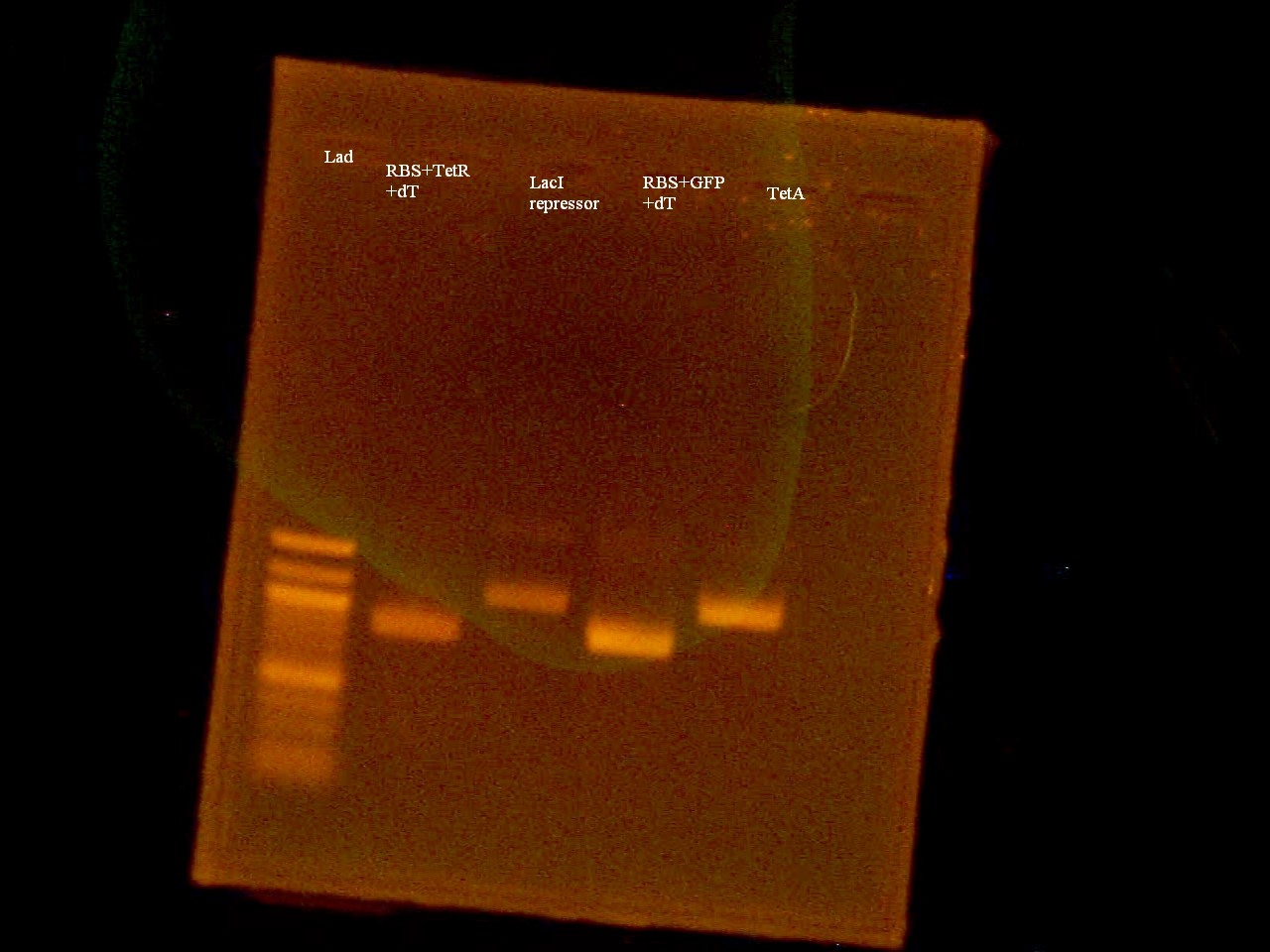
Ran products from plasmid prep (pSB1A7 x 4 samples) on 1% agarose gel for 27 minutes.
-Lane 1 - 1 kb GeneRuler ladder (2 uL) -Lane 2 -6 pSB1A7 (3 uL) + 6x loading dye (2 uL); Mixed up what sample was in Lane 4, so Lanes 5 and 6 were run.
Conclusion: The bands appeared to be at the correct size for pSB1A7.
August 22, 2008
Christa, Munima, Nathan Puhl, Roxanne
Objective: Run a gel to confirm that appropriate inserts were amplified from the PCRs and do a gel extraction of the inserts to prepare them for the biobrick format.
-Could not obtain a picture of the gel (1% agarose) of the half of the digested pSB1A7 (15 uL x 3 wells) and the recently amplified CheZ gene (15 uL x 3 wells) because the camera would not turn on. The CheZ gene appeared at the correct size (~700 bp). -Did a gel extraction of pSB1A7 and CheZ from that gel with the Qiagen MiniElute Gel Extraction kit. Final volume of each was 10 uL. -Ran another 1% agarose gel to confirm that the gel extraction was successful. Ran 1 uL of each sample. Bands appeared at appropriate sizes. The concentrations of pSB1A7 and CheZ were estimated to be 25 ng/uL and 80 ng/uL, respectively.
Next step: Digest pSB1A7 with antarctic phosphatase. A restriction digest will be performed at a later date on CheZ to prepare it for insertion.
August 26, 2008
Roxanne, John
-Restriction Digested the Biobrick Parts I13504, I13401, P0440, C0014, B0015, J31007 with XbaI and PstI
-20 uL template (~2 ug) -5 uL React 2 -4 uL XbaI -4 uL PstI -17 uL ddH2O ____ 50uL Reaction in 37.0C H2O bath overnight
August 27, 2008
Roxanne
-Inactivated the Restriction Enzymes by placing them on the heating block at 65.0C for 10 minutes -Ran 25uL of DNA on a 1% Agarose Gel at 100 V for 30 minutes -Gel Extracted the DNA -Ran 1 uL of DNA in a 1% Agarose Gel at 100V for 30 minutes.
August 28, 2008
Roxanne, Munima, Sebastian, Nathan Puhl
-Performed a restriction digest on LacI+dt, pLacI, pStrong, and RFP sub
-10 uL template -5 uL NEB 2 -2 uL Restriction Enzyme #1 (Xba I or Spe I) -2 uL Restriction Enzyme #2 (Pst I) -21 uL ddH20 ____ 50 uL Rxn left to run overnight at 37.0
-Ran a Gel of the Riboswitch PCR which had been done with Taq a couple of weeks ago to quantify the amount of DNA present. Setup the Math to perform a ligation into pGEM T-easy.
-Picked a colony from the dT plate which was stored in the fridge from several weeks ago, incubated overnight at 37.0C
August 29, 2008
Roxanne, Nathan Puhl, Munima, Sebastian, Andrew
-Performed the ligation of the riboswitch into pGEM T-easy, transformed and plated.
=Roxanne
-Gel Extracted the remainder of the DNA
August 30, 2008
Nathan Puhl, Roxanne, Andrew
-Ran a gel of the Digested Parts on 1% Agarose @ 100V for 27 minutes.
 "
"