Team:University of Lethbridge/Project
From 2008.igem.org
The “Bacuum" Cleaner – an intelligent self-propelling keener cleaner
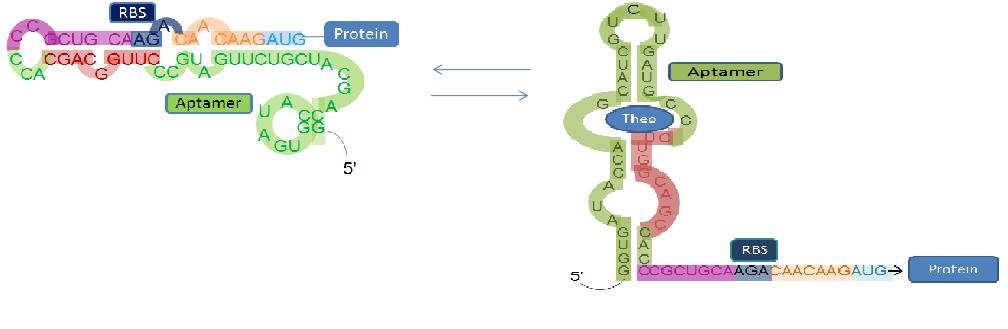
Our goal is to create a modified Escherichia coli bacterium capable of seeking out and degrading toxic aromatic pollutants created during the oil refinery and mining processes. Our “Bacuum" cleaner will respond to this destructive compound through interaction with a programmed riboswitch (an mRNA capable of binding a target molecule with its 5’ UTR which causes a conformational change, thereby regulating any downstream protein translation). By using riboswitches that switch at varying concentrations of target ligand, we can alter the signal which is induced. At low concentrations, we intend to have our riboswitch express the motility protein cheZ in E. coli, thus directing the bacterium towards higher concentrations of our target molecule (i.e. positive chemotaxis). Once it reaches a threshold concentration, a catabolic pathway capable of degrading our target pollutant will be activated.
We plan to use [http://en.wikipedia.org/wiki/Systematic_Evolution_of_Ligands_by_Exponential_Enrichment SELEX] to reprogram the theophylline riboswitch initially characterized by our team last year. For our proof-of-principle concept, we chose 2-chlorobenzoate, a compound related to polychlorinated biphenyls (PCBs), as our target molecule. This not only builds on our past work with riboswitches but also demonstrates an alternative function for riboswitch-controlled chemotaxis, changing from a simple detection to active “search and destroy” role. Fluorescent protein expression will be used to demonstrate and characterize the functionality of the riboswitch.
Our project also has major environmental implications, especially in Alberta, where the oil industry is the driving economic sector. The tailings ponds used to house discarded mining refuse from the oil refineries pose a major environmental dilemma. The problem arises with being unable to collect and degrade the compounds which make up these toxic soups. These not only toxic but often corrosive water beds not only affect the local environment but can also cause severe repercussions to other ecosystems as much of the wildlife relies on getting their food and water from the now disrupted bionetwork. These tailings ponds are often laced with heavy metals and dangerous chemicals, including these toxic aromatic hydrocarbons which can be extremely difficult to breakdown. Despite the efforts of the corporations responsible, some of these chemicals can leech into and contaminate the surrounding ground water and soil. It is our intention that our “Bacuum” will not only remove these compounds from such an aqueous system, but will also eliminate them. This goes one step further than the ordinary less-intelligent bag-dependent vacuum cleaners. Ultimately we aim to have our bacteria convert the target aromatic into a useable compound. One example would be the biosynthesis of fatty acids as an alternative fuel source, creating a potential “Bioreactor” which is capable of creating a more environmentally friendly fuel source.
Project Details
 Chemotaxis - the "search" from "search and destroy"
Chemotaxis - the "search" from "search and destroy"
Our subproject is to ensure our 'bacuum cleaner' will search out and move towards our potential target. Using a riboswitch which responds to this target, a gene involved in motility in Escherichia coli (CheZ) will be switched to an 'on' state (Topp and Gallivan, 2007). CheZ controls bacterial movement by dephosphorylating CheY, altering the bacteria from random tumbling to directed movement. By doing so, we hope to observe the bacteria moving towards the target molecule in a positive chemotaxis manner.
Because we wish to eventually have our bacteria degrade this activating compound, we need to have a multi-level response system established. Our bacteria should respond to low levels of the target by activating our CheZ riboswitch (through a strong binding aptamer) and to high levels by activating the catabolic pathway responsible for its degradation. By doing so, we will have created our 'search and destroy' system.
Our subgroup will attempt to establish a working motility assay to prove control of chemotaxis is possible with an already characterized theophylline-binding riboswitch and later with our new target molecule. We will also use a colour read-out system of two fluorescent proteins to demonstrate the differential binding of the ligand at varying concentrations.
 Riboswitch Characterization
Riboswitch Characterization
This subproject was created to quantify and characterize the binding affinity of the theophylline riboswitch to theophylline as well as other similar aromatic ligands. We will use Green Fluorescent Protein (GFP) sequenced behind the riboswitch to help in this endeavour. When the target ligand is bound to the riboswitch, the production of GFP will be induced, producing green colonies. When the ligand is not bound, GFP will not be produced. We intend to try to characterize the riboswitch with and without the iGEM scar to determine if this produces any effect on the relative effeciency of the riboswitch on the protein.
We will use variable concentrations of Theophylline, Caffeine and 3-methylxanthine to help with the characterization of the initial theophylline riboswitch. We will also test other aromatic compounds to determine wether or not, the theophylline riboswitch can be utilized to bind them as well.
Once we have successfully produced mutant riboswitches through SELEX, will we then perform these same characterization experiments to determine their binding affinities.
SELEX
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) is a method used to produce novel nucleic acid sequences, referred to as aptamers, that bind specific ligands. SELEX involves screening a library of possible aptamers for binding capability repeatedly until a sequence of the desired specificity or binding affinity is found.
SELEX will be used in our project to produce multiple riboswitches capable of specifically binding the target toxin at varying concentrations. Multiple riboswitches are needed to allow a separation between the "search" and the "destroy" functions of our Bacuum cleaner, with the CheZ search ability controlled at low ligand levels and the degradation pathway controlled at high levels.
 TIR - our unique "watermark" and a form of detection
TIR - our unique "watermark" and a form of detection
Translation initiation regions (TIRs) are elements found in certain eukaryotic mRNAs that facilitate translation initiation by noncanonical, end independent interactions (Hellen, 2007; Skorski et al., 2006). We are going to incorporate the TIR from the rpsA gene, encoding ribosomal protein S1, of E. coli into our "Bacuum cleaner" with green fluorescent protein as a means of monitoring the location of our cells using minimal cell resources.
The TIR from the rpsA gene will be characterized through the comparison of translation rates for GFP to other canonical RBS. This will allow us to standardize the number of ribosomal initiations per second (RiPS). In generating these parameters we will be able to contribute to building a database of standard parts that can be used to generate biological systems that have a greater degree of reliability.
 Degradation of 2-chlorobenzoate
Degradation of 2-chlorobenzoate
Polychlorinated biphenyls (PCBs) have been established as toxic and environmentally unfriendly since the early 1980s. One potential intermediate step in the degradation of PCBs involves elimination of the chloro substituents. A dehalogenation pathway for ortho-halobenzoates was discovered in Pseudomonas aeruginosa strain 142 and successfully isolated and characterized by Tsoi et al. (1999). This metabolic pathway, comprising of the ohb genes, catalyzes the degradation of 2-chlorobenzoate into catechol. We aim to combine this system with the catechol 2,3 dioxygenase xylE gene (Ingram et al., 1989) to further break down catechol into the highly-coloured 2-hydroxymuconic semialdehyde. By doing so, we will combine our degradation system with a reporter system, bringing yet another level of intelligence to our incredibly awesome “bacuum cleaner.”
References
Hellen, C. U. T. 2007. Bypassing translation initiation. Structure 15, 4-6.
Ingram, C., Brawner, M., Youngman, P., and Westpheling, J. 1989. xylE functions as an efficient reporter gene in Streptomyces spp.: Use for the study of galPI, a catabolite-controlled promoter. J. Bacteriol. 171, 6617-6624.
Jenison, R. D., Gill, S. C., Pardi, A., and Polinsky, B. 1994. High resolution molecular discrimination by RNA. Science 263, 1425-1429.
Lynch, S. A., Desai, S. K., Sajja, H. K., and Gallivan J. P. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 14, 173-184.
Nomura, Y. and Yokobayashi, Y. Dual selection of a genetic switch by a single selection marker. BioSystems 90, 115-120.
Skorski, P., Leroy, P., Fayet, O., Dreyfus, M., and Denmat, S. H. 2006. The highly efficient translation initiation region from the Escherichia coli rpsA gene lacks a Shine Delgarno element. J. Bacteriol. 188, 6277-6285.
Topp, S. and Gallivan, J. 2007. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 129, 6807-6811.
Tsoi, T. V., Plotnikova, E. G., Cole, J. R., Guerin, W. F., Bagdasarian, M. and Tiedje, J. M. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65, 2151-2161.
 "
"

 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]