Team:University of Sheffield/Rosie
From 2008.igem.org
| Introduction | Our project | Modelling | Wet Lab | Our team | Timetable | Miscellaneous |
|---|
| Introduction | Protocols |
|---|
Rosie's Lab Book
Contents |
Early Work
Training, involving being familiarised with all the protocols listed with extensive practising. Some pHD students had some free labour at this point :)
Our Project Experiments
Our first PCR to produce the BarA homology sequences on kanamycin cassette did not work. After varying magnesium concentrations from 0.5mM to 4.5mM proved successful for the 3.5mM concentration.
We repeated the PCR with Phusion polymerase, which has a much lower rate of error than Taq.
Now we started to electroporate.
We stated by making our MG1655's electrochemically competant and glycerol stocking them.
Electroporations 1 - 5 failed. Various troubleshooting, varying conditions and concentrations of most relevant factors. Eg 100ul competant cells rather than 40ul. Many different DNA concentrations. Recovery time increased 3fold.
Positive controls failed on cells known to competant (our advisor gave us them and tested them previously). After 4th electroporation we tested the ampicillin which had been opened brand new when we started using is. Turned out to be faulty. Ordered new ampicillin.
Were given a strain of E.coli with BarA alreadt knocked out by the Karolynska institute in Sweden. Great! Problem was their knockout method had left the strain both ampR and kanR. pKD46 is also ampR and kanR. So we tried to modify pKD46 to make it chmR instead.
To make pKD46 chmR, we wanted to restrict out kanR from pKD46 and chmR from pACYC (a plasmid provided by another pHD student). However, the only restriction enzyme suitable for pKD46 in this position was Mlu1, which we borrowed from out department. However, we were told it was old and may not work. The restriction didn't work, and this may be the reason.
Biobrick Book Testing
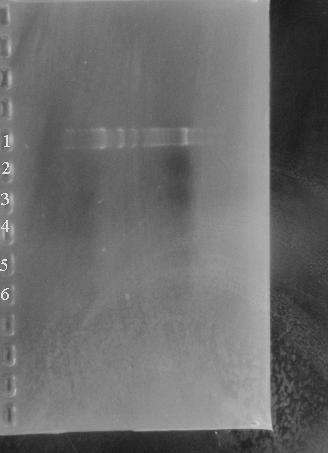
After Eva's characterisation failed, we had our suspicions about the BioBrick Registry. However, given our own intermediate skill, and the prescence of many far more likely problems, it wasnt until we started to make the BioBricks that the full scale of the problem was obvious. After extractng and restriction the BioBrick vector pSB10A3, nothing showed up on the gel.
Lane1: Marker
Lanes 2 - 6: BBpSB1AK3 digested
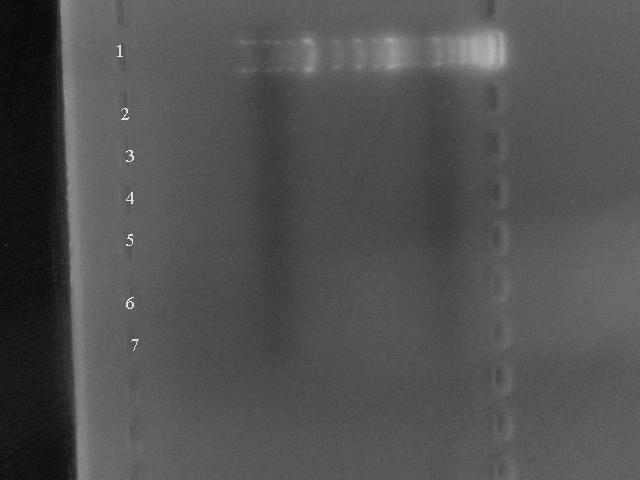
Below is the gel of 5 extractions from BioBricks all over the registry (emphasis on vectors as thats waht we needed) - our final test to see if the book was faulty or not:
Lane 1: Marker
Lane 2: pSB1AK3
Lane 3: pSB1AC3 source plate:1020 well:4B
Lane 4: pSB1A2 sp:1003 w:4A
Lane 5: pBAD/araC sp:1013 w:5C
Lane 6: 30C6HSL reciever sp:1001 w:12B
Lane 7: Banana odour sp:1021 w:12B
As you can see, there is no DNA present.
 "
"