Team:Wisconsin/Modeling
From 2008.igem.org
(→Computer Modeling) |
|||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html><link rel="stylesheet" href="https://mywebspace.wisc.edu/jluo1/wisconsinstyle.css" type="text/css"></html> | ||
| + | |||
<div style="background: #000; padding: 10px;"> | <div style="background: #000; padding: 10px;"> | ||
| - | <div style="width:800px; height:147px; border:0px; margin:0px;">[[Image:Igemwibanner. | + | <div style="width:800px; height:147px; border:0px; margin:0px;">[[Image:Igemwibanner.gif]]</div> |
| + | __NOTOC__ | ||
| + | <div class="NavBar"> | ||
| + | {| | ||
| + | !align="center"|[[Team:Wisconsin|Home]] | ||
| + | !align="center"|[[Team:Wisconsin/Team|The Team]] | ||
| + | !align="center"|[[Team:Wisconsin/Project|The Project]] | ||
| + | !align="center"|[[Team:Wisconsin/Parts|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Wisconsin/Modeling|Modeling]] | ||
| + | !align="center"|[[Team:Wisconsin/Notebook|Notebook]] | ||
| + | |}<br> | ||
| + | </div> | ||
| - | + | <div class="Main"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Computer Modeling== | ||
| + | <br><font color=#aada84> | ||
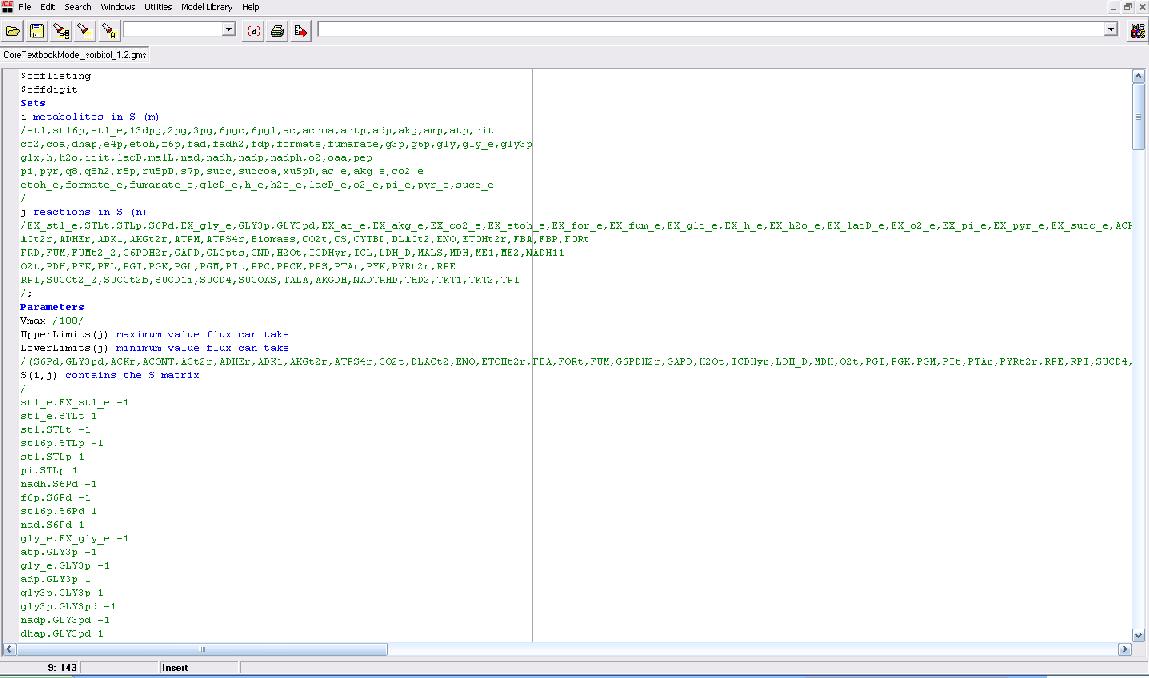
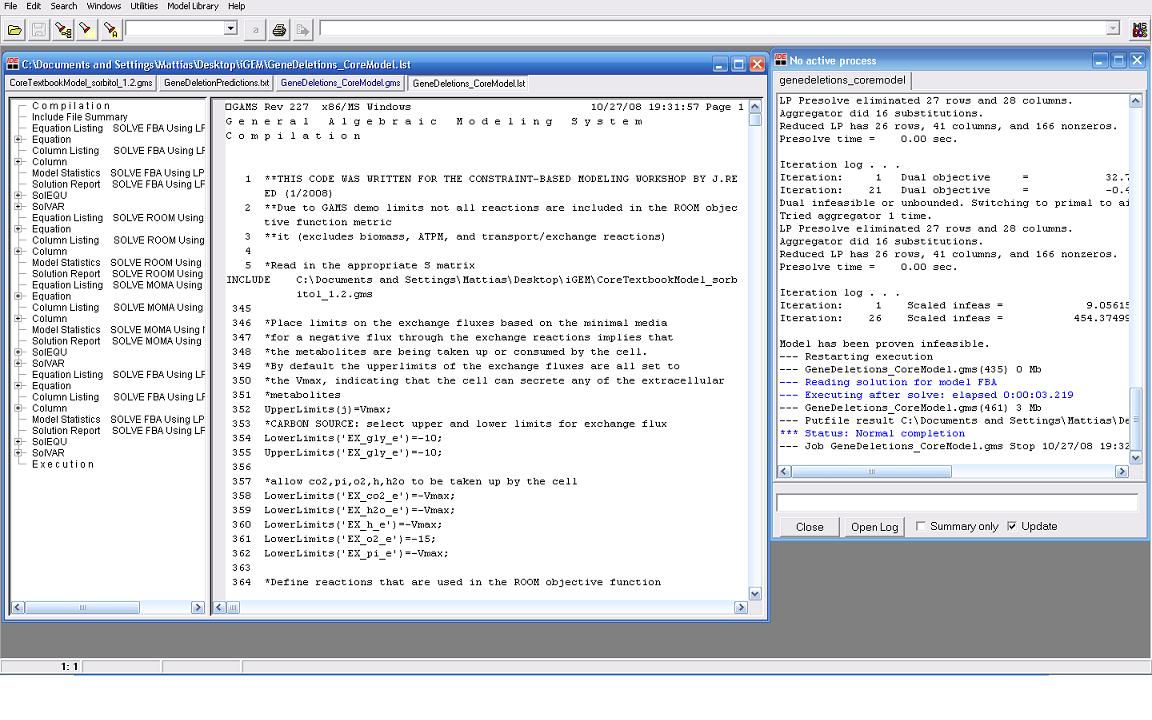
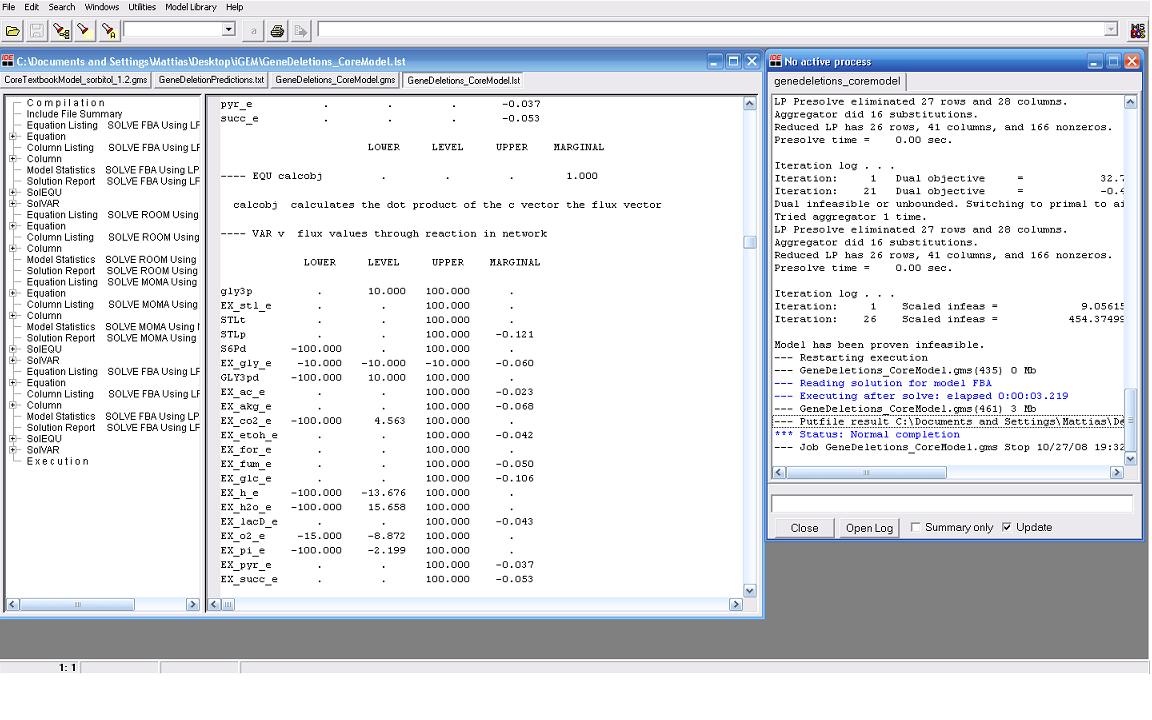
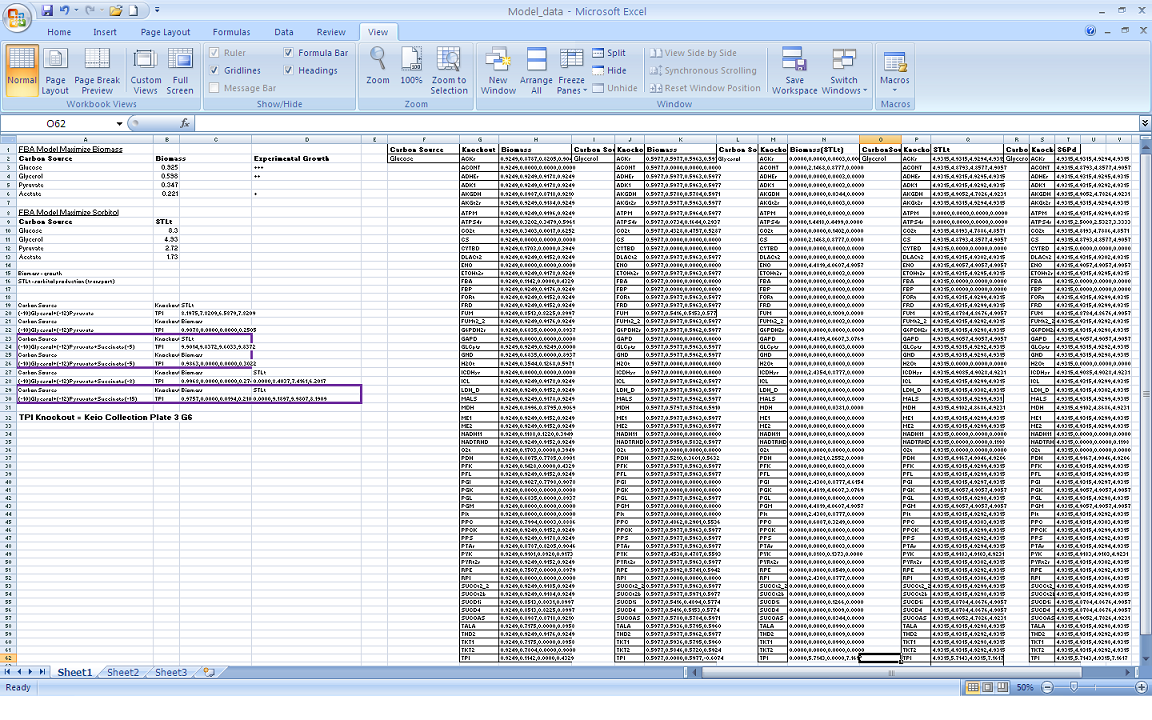
| + | Our modeling adventure started with a brief introduction to programming using GAMS software and an ''E. coli'' database developed by Dr. Jennie Reed at the University of Wisconsin College of Engineering. Dr. Reed’s database works in conjunction with the GAMS software to measure metabolic fluxes in ''E. coli''. Using this as a base point we were able to computationally manipulate metabolites, genes, and the growth environment to determine the most optimal conditions for cell growth and hopefully sorbitol production. Glycerol is currently produced at a rate of one pound for every ten pounds of biodiesel produced. Consequently, our team felt it would be interesting if we could find a novel use for glycerol in our pursuit of sorbitol producing ''E. coli''. As such we decided to utilize glycerol as our growth media carbon source. After many hours spent running simulations we identified several potential candidates which we are currently working with. | ||
| - | + | [[Image:Printscreen.JPG|600px]]<br><br> | |
| - | + | [[Image:Printscreen_2.JPG|600px]]<br><br> | |
| - | + | [[Image:Printscreen_3.JPG|600px]]<br><br> | |
| - | + | [[Image:Printscreen_4.JPG|600px]]<br><br> | |
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | | | + | |
| - | < | + | |
Latest revision as of 00:27, 30 October 2008
Computer Modeling
Our modeling adventure started with a brief introduction to programming using GAMS software and an E. coli database developed by Dr. Jennie Reed at the University of Wisconsin College of Engineering. Dr. Reed’s database works in conjunction with the GAMS software to measure metabolic fluxes in E. coli. Using this as a base point we were able to computationally manipulate metabolites, genes, and the growth environment to determine the most optimal conditions for cell growth and hopefully sorbitol production. Glycerol is currently produced at a rate of one pound for every ten pounds of biodiesel produced. Consequently, our team felt it would be interesting if we could find a novel use for glycerol in our pursuit of sorbitol producing E. coli. As such we decided to utilize glycerol as our growth media carbon source. After many hours spent running simulations we identified several potential candidates which we are currently working with.
 "
"