Template:Team:UC Berkeley/Notebook/MJV notes
From 2008.igem.org
(→Madhvi 20:17, 25 September 2008 (UTC)) |
(→Madhvi 22:39, 30 August 2008 (UTC)) |
||
| Line 394: | Line 394: | ||
# Zymo cleanup by adding 300uL of ADB buffer and doing two washes with 200uL each. Elute with 6uL of water. | # Zymo cleanup by adding 300uL of ADB buffer and doing two washes with 200uL each. Elute with 6uL of water. | ||
# Ligate with 1uL EcoRV digest, 1uL Lucigen Fosmid blunt vector, 1uL Ligation Buffer, 6.5uL water, and 0.5uL T4 DNA ligase. Incubate at room temperature for 30 minutes. | # Ligate with 1uL EcoRV digest, 1uL Lucigen Fosmid blunt vector, 1uL Ligation Buffer, 6.5uL water, and 0.5uL T4 DNA ligase. Incubate at room temperature for 30 minutes. | ||
| - | # Add one aliquot of epicentre TrfA cells | + | # Add one aliquot of epicentre TrfA cells to ligation and leave on ice for 30 minutes. |
# Heat shock for 30 seconds at 42C. | # Heat shock for 30 seconds at 42C. | ||
# Incubate 2 minutes on ice and then rescue with 100uL 2YT and 0.2uL of inducer 1000X. | # Incubate 2 minutes on ice and then rescue with 100uL 2YT and 0.2uL of inducer 1000X. | ||
Latest revision as of 21:00, 25 September 2008
Madhvi 05:43, 4 June 2008 (UTC)
Yesterday we talked about biobricks and cloning. Today we talked about assembly, BBb standard, and documentation. We also discussed our project plan today. In our demos, we run a PCR yesterday to amplify GFP from a plasmid. Today, we did a zymo cleanup, a restriction digest, and ran an analytical gel (E-gel).
I set up my notebook today and I am looking at papers to try to find a good choice of protein for the protein purification part of the project. I am also trying to find growth-phase dependent promoters.
Madhvi 05:08, 12 June 2008 (UTC)
Last week, I didn't really find anything regarding a good choice of protein, but I did find the rrnB P1 promoter that robustly turns off when cells enter the stationary phase. I also designed the oligos for my basic parts (variations of Cre, ligase, BglII, BamHI and the rrnB P1 promoter). Construction files for these parts (K112100-K112121) are included in the construction files section of my notebook. All of my oligos have not yet come in. They should be in tomorrow and I should be able to run PCRs tomorrow.
In the meantime, I have been doing a few things to determine the assay conditions for the Gateway reactions. On Friday and Monday, Jin and I made concentrated stocks for the six parts of the Gateway buffer (1M Tris pH 7.4, 5M NaCl, 1M Spermidine, 0.5M EDTA pH 8.1, 50mM DTT, 10 mg/mL BSA). Yesterday, I mixed appropriate volumes of them to make 10mL of a 1x Gateway Buffer (22mM Tris, 100mM NaCl, 7mM Spermidine, 5mM EDTA, 2mM DTT, 1mg/mL BSA) and stored it at 4 degrees. Jin says it should be stable for approximately one week.
Today, Jin and I ran the regular clonase reaction with pBjh1600 (RFP entry vector with p15a origin) and pBjh1601CK (CK assembly vector with p15a origin). The reaction is set up with 0.6uL EB Buffer (Dilute Tris), 0.3uL Assembly vector miniprep, 0.3uL Entry vector miniprep, and 0.3uL LR Clonase II. We also ran this reaction (usually 1.5uL) scaled up to 10uL by adding 8.5uL of 1x gateway buffer. We transformed both products into DH10B and plated on KC plates after 1hr. reaction time. We will compare the number of colonies on both plates tomorrow.
I also am growing up cultures two cultures which we will use tomorrow for a Gateway reaction after arabinose induced lysis. The first culture is TG1 (ccdB immune strand) with pSB3C6-Bjh1311 (holin, antiholin and lysozyme under Pbad promoter; p15a origin), pHA57 (xis, int; high copy number, temp. sensitive origin of replication), and pBjh1600. The second culture is TG1 with pSB3C6-Bjh1311, pHA57, and pBjh1601AK. We will mix given amounts of the two culture, wash and resuspend in our 1x gateway buffer and then lyse with arabinose. We will transform the resulting reaction mixture into DH10B and if the reaction was successful we should see colonies on AK plates.
I am also growing up cultures of Righty and Lefty cells to make competent cells that I can use to make my BamHI and BglII basic parts.
Madhvi 08:02, 17 June 2008 (UTC)
Thursday, I made Righty and Lefty competent cells. When I checked the plates from the Gateway reactions that we ran, I saw only a couple of colonies on the plate where the reaction was scaled up and a lot of colonies on the other plate. So, it looks like the reaction does not really work when scaled up to 10uL. Chris thinks that we need to find more information about the appropriate buffer solution and we need to screen different potential buffer solutions to find one that works.
My oligos also came in on Thursday, so I started running PCRs. Because we couldn't find the templates for the BglII, Cre and BamHI experiments, I ran the PCRs on MG1655 and T4 genomic DNA to clone the rrnB P1 promoter and parts of ligase (used program 55). However, none of the reactions worked. So, on Friday, I tried running the MG1655 PCR with & without DMSO as well as the ligase PCRs and the BamHI PCRs without DMSO (on program 55). The only thing that worked was the MG1655 PCR with DMSO. On Friday, I also transformed appropriate templates into Righty and Lefty cells [pBca9145-Bca1010 (Cre) into Righty, pBca9145-I716210 (BamHI) into Righty and BglII with a promoter into Lefty]. I also did test transformations of BamHI and BglII into the Righty and Leffty competent cells that I made in order to confirm that they were correct (which they were).
On Saturday, I grew up colonies from the plates that I made in order to be able to isolate template. However, the Amp media that I used appeared to be lethal, so I did not have cultures on Sunday. I grew up cultures again on Sunday in new Amp media and was able to miniprep and run PCRs on the Cre and BglII templates today.
In the meantime, did PCRs on the BamHI and ligase parts on Saturday with DMSO at 55. I succeeded in amplifying the first three segments of ligase and in getting products for {r.BamHI!}, {r.BamHI>}, and {<r.BamHI>}. Since the bands for the BamHI parts on the gel were quite faint, I used them as templates on Sunday to amplify them more (I ran these reactions at 55 with DMSO). I also did a SOEing reaction with the first two segments of ligase at 55 with DMSO. I tried to clone the {a~r.BamHI!} and {rbs.r.BamHI!} parts using {r.BamHI!} as a template by running these reactions with & without DMSO at 45. I also tried cloning the 4th fragment on ligase with and without DMSO at 45. I found the the {r.BamHI!} template was impure so the resulting reactions had a lot of background and the products were difficult to resolve on the gel. However, the {r.BamHI>} and {<r.BamHI>} reactions looked good and gave well-defined bright bands. Furthermore, the 4th segment of ligase was successfully amplified both with and without DMSO. The SOEing reaction with the first two segments of ligase was also successful.
Today, I reran the {r.BamHI!} (55 w/ DMSO), {a~r.BamHI!}, and {rbs.r.BamHI!} (both 45 with and without DMSO) PCRs using {r.BamHI>} as a template. I also ran the SOEing reaction with the third and fourth segments of ligase with DMSO at 55). I also ran all of the BglII and Cre reactions with the templates that I miniprepped from my cultures.
Madhvi 03:02, 19 June 2008 (UTC)
All of the PCRs worked except for the SOEing reaction of the third and fourth segments of ligase. So, I went ahead with the next cloning steps (digestion, ligation, and transformation) yesterday. Today, I got colonies on all of my plates except the {r.BamHI>} plate. I will redo the ligation & transformation for this one sample tomorrow. I grew up colonies from all of the other plates today, so that I can miniprep and send them in for sequencing tomorrow. Since the SOEing reaction for the third and fourth segments of ligase didn't work yesterday, I reamplified the third region of ligase and segment E (combination of first and second regions) in a PCR that Christie set up for me yesterday evening. I used the reamplified part C to run SOEing reactions of C & D at both 45 & 55 with and without DMSO. The SOEing reaction worked at 55 without DMSO, so I used the resulting fragment (F) as a template along with fragment E to run the final SOEing reaction for ligase. I will see how it works tomorrow!
Madhvi 02:12, 20 June 2008 (UTC)
The ligase PCR worked with 2K45 w/o DMSO! yay! So, now that gave me the {rbs.ligase!} part. I digested that today and ligated it with the new stock of the digested parent vector that Molly, Aron, and Bing made and purified. I also ligated {r.BamHI>} because that plate had no colonies on it yesterday. I transformed both of the samples into Righty cells and plated. Bing miniprepped all of the samples that I grew up yesterday and sent them out for sequencing.
Jin found another paper that described a slightly different buffer mix for the gateway reaction. We will test it tomorrow using the two strains that have the arabinose induced lysis device, along with an entry/assembly vector, and the vector to make xis and int (TG1 with pSB3C6-Bjh1311, pHA57, pBjh1600 and TG1 with pSB3C6-Bjh1311, pHA57, pBjh1601AK). I grew up these two strains in ACSpec and ACK media, respectively.
Today, Chris presented to us about Adobe Illustrator and I met with Susanna Spiro (from COE), Marlee, and Nade about the blog today. We should be starting that next week.
Madhvi 08:10, 23 June 2008 (UTC)
Friday was a long and eventful day. I got about 20-25 colonies on the {rbs.ligase!} plate and 8 colonies on the {r.BamHI!} plate. Chris helped me run a colony PCR on 8 of the ligase colonies to screen them and make sure that the part that they had was the right size. Basically, each colony was picked swirled around in the appropriate PCR tube then put into a well with LB on a 24 well block. The PCR was run with the standard forward and reverse oligos for BBb Biobrick parts (ca998 and g00101). Out of the 8 clones screened, clone #2 had the wrong part (it was ~1000bp instead of ~1500bp). All of the other clones appeared to be correct on the gel that was run.
I also got back sequencing results. All five of the BglII parts were perfect. All five of the Cre parts were perfect partials--I told Jin to ask them to sequence all five of them with the standard reverse oligo (g00101) so that I can check the last 300bp of each of the Cre parts. The rrnB P1 promoter had a perfect sequence. Out of the four BamHI parts that were sequenced, two of them ({r.BamHI!} and {rbs.r.BamHI!}) were bad reads. {a~r.BamHI!} had a huge deletion in the middle of the part. {<r.BamHI>} had a point mutation, which was not silent. I re-picked colonies for all of the BamHI parts on Friday so that they could be miniprepped and sent for sequencing on Saturday. Jin suspected that the BamHI template that I used (pBca9145-Bjh0029) was bad, so I also set up PCRs using pBca9145-I716210 as template for all of the BamHI parts. I went ahead and set them up at 55 w/ & w/o DMSO and at 45 w/ & w/o DMSO.
On Friday, Jin and I also screened 15 buffers using the cultures that I grew up on Thursday for the Gateway reaction with arabinose-induced lysis. Christie used the stock solutions to make 10mL of the new variation of 1x Gateway buffer (25mM Tris, 6mM Spermidine, 5mM EDTA, 82mM NaCl, 0.5 mg/mL BSA, 2.5mM DTT). The buffers that we screened were: the 2 variations of the Gateway buffer that we had made, NEB1, NEB2, NEB3, NEB4, Expand 2, Expand 3, Phusion GC, Phusion HF, & Fast Digest. Chris had screened many of these by scaling up the clonase reaction, but he got inconclusive results (many of them had both red and white colonies) because the clonase that we have has probably gone bad.
The protocol that we used for the experiment involved doing the following for each sample:
- Mix 300uL of each culture in Eppendorf
- Centrifuge using maximum speed for 30 sec
- Discard supernatent & remove residual liquid using pipette
- Resuspend in 100uL of 1x buffer
- Centrifuge using maximum speed for 30 sec
- Discard supernatent & remove residual liquid using pipette
- Resuspend in 100uL of 1x buffer
- Add 1uL of 20% arabinose
- Incubate at 37 degrees for 1 hour
- Incubate for 1 hour at room temperature
- Centrifuge using maximum speed for 30 sec
- Transform 10uL of the supernatent into 45uL of Righty cells
- Rescue with 50uL of 2YT and incubate for 1 hour at 37 degrees
- Plate on AKG plates (Righty cells have Gentamycin resistance)
After incubating 1 hour at 37 degrees with arabinose, the samples in Expand 3, Gateway Buffer #1 (the one I made originally) and Phusion HF showed apparent lysis (they were viscous when tested with a pipette). Anyway, none of the plates had colonies on them on Saturday. Jin feels that there might be something wrong with the expression of xis and int from the cells, which would explain why none of the reactions worked.
On Saturday, I found that the PCRs for the BamHI parts all worked at 55 w/o DMSO. So, I went ahead and digested, ligated, and transformed all of them BamHI parts into Righty cells. Jin and I also miniprepped the cultures that were grown up on Friday (including my ligase cultures, my BamHI cultures, and some cultures for Dirk and Bing). I also grew up colonies for Aron and Molly from their plates.
Today, I found out that 2 out of the five BamHI parts ({r.BamHI>} and {rbs.r.BamHI!}) that I plated had no colonies and {a~r.BamHI!} had only one colony). Jin thinks that the vector digest that I used was too dilute. I will redo the ligation and transformation steps for those parts tomorrow. I also got sequencing back from yesterday's samples. Ligase #5 is perfect! yay! So, I went in and set up PCRs for the rest of the ligase parts (the one that I have right now if {rbs.ligase!}). I set them up at 2K55 w/ & w/o DMSO and at 2K45 w/ & w/o DMSO. With the exception of {<r.BamHI>}, the other BamIi parts seem to be problematic (either mutations or bad reads again). So, I guess that I will follow through with the cloning from I716210 for the other BamHI parts and see how it goes. I will check the reverse sequences for the Cre parts tomorrow.
Madhvi 06:00, 26 June 2008 (UTC)
On Monday, I analyzed the reverse reads of my Cre sequences. Two of them were bad reads, two of them appeared to be the wrong parts and one of them was correct. Jin sent the four that appeared to be bad for resequencing on Monday and they all were correct in the reads that I got on Tuesday. So, all of the Cre parts that I have made thus far are correct.
On Monday, I also PCRed the {<r.BglII!} and {Cre!} parts because we realized that templates that we had used to clone them were not in Biobricks, so an Eco/Bam transfer would not work for these. I digested the PCR products of this. I also digested the five ligase parts that I PCRed from the {rbs.ligase!} part (all of the PCRs worked well at 55 w/o DMSO). Bing did the ligations and transformations for these ligase parts, the {Cre!} part, the {<r.BglII!} part and for the three BamHI parts that had no colonies or only one colony on the plate on Sunday ({r.BamHI>}, {a~r.BamHI1}, and {rbs.r.BamHI!}). I forgot to tell him to transform the BamHI parts and the into Righty cells and the BglII part into Lefty cells, so he put them all in DH10B. The {<r.BglII!} part works just fine in DH10B cells. He got the labeling for the BamHI plates mixed up, so I am not sure which one is which. Two of the plates had a few colonies and the other one had no colonies. I suspect that the one with no colonies is the {rbs.r.BamHI!} part.
Yesterday, Bing grew up samples from all of the plates from Monday (the 5 ligase parts, the {Cre!} part, the {<r.BglII!} part, and the two BamHI plates that had colonies). He and Molly miniprepped them roday and sent them out for sequencing. I also redid the ligations and transformations of the three BamHI parts ({r.BamHI>}, {a~r.BamHI1}, and {rbs.r.BamHI!})into Righty cells yesterday. However, today I found that these plates had very few colonies (a total of 13 for all three plates). I did colony PCR on 11 of the colonies today and found that only one of them appeared to be the right size. Tomorrow, I will miniprep and send this colony for sequencing; however, I suspect that it is probably not right. Jin thinks that there might have been an accidental tube switch in the process of making these competent cells and these cells might actually be Lefty cells. Today, I asked Sherine and Cici to redo the ligation and transformation of the three BamHI parts (this time I made sure that they transformed into Righty cells).
Yesterday, I also transformed minipreps of all of the BglII parts that I have, the {<r.BamHI>} part, the rrnB P1 promoter, and the {rbs.ligase!} part into DH10B (because they were all made in Lefty or Righty cells and had the BamHI or BglII sites methylated). The BglII/BamHI samples that can survive in DH10B are {<r.BamHI>}, {r.BglII>}, and {<r.BglII>}. Today, I picked colonies from these plates and the rrnB and ligase plates to be miniprepped tomorrow so that we can have plasmids with free BglII and BamHI sites for assembly. From Monday, we also know that the {r.BglII!} part is fine in DH10B. All of them gave hundreds of colonies with the exception of {r.BglII>}, which seems relatively toxic compared to the other three parts.
With regards to Gateway stuff, Chris told Jin to order a small size of Clonase so that we can repeat the experiment where we scale up the reaction with different buffers. We also are planning to do a couple of other experiments. The possible explanations for the failure of Friday's experiment are that the xis and int did not have enough time to catalyze a significant number of transfers (we only left them 1hr to do Gateway, which works for clonase, but maybe the xis and int are being produced at too low concentrations for it to work in 1hr). It also may be a case where they are not catalyzing enough transfers for them to be assayable (the pAH57 plasmid has been used before to park things in the genome via recombination, but this requires only a single transfer). So, we are going to see whether there is an assayable amount of transfers that occur in the cytoplasm over a longer time scale. We are planning to put pAH57 (xis & int cassette; temp. sensitive origin; AmpR), pBca1256 (entry vector; pUC origin; SpecR), and pBjh1601KC (assembly vector; p15a origin; KanR & CamR) into TG1. After growing overnight, we will then miniprep the mixture of plasmids and transform into DH10B and plate on KC plates.
Chris also suggested testing the effect of the xis/int construct on the growth rate of TG1 (they are somewhat toxic, so they have shown slow growth in the past). We are making saturated cultures of the following things and then doing the same dilution on all of them and testing OD after a set amount of time:
- TG1
- TG1 with pAH57
- TG1 with pAH57 & pBca1256
- TG1 with pAH57 & pBjh1601KC
- TG1 with pAh57, pBca1256, & pBjh1601KC
Chris thinks that the toxicity effect might be greater if there are substrates of the xis and int present in the cytoplasm of the cell, but we'll see what happens when we actually do the experiment.
For the Gateway stuff, I transformed pAH57 into TG1 competent cells and plated on Amp plates. Today, I grew up colonies and I transformed either pBca1256 or pBjh1601KC into them when they were at midlog using the protocol posted for transforming into cultures.
Madhvi 08:57, 27 June 2008 (UTC)
The transformation of the BamHI parts from yesterday did not really work again--there was only one colony between all of the three plates. Chris suspects that the toxicity of these parts is leading to problems with the transformation. Today, I did an EcoRI/BamHI digest of the pBjh1600 plasmid (entry vector with p15a origin) and gel purified. I did new PCRs of all of the BamHI parts except {<r.BamHI>}, which sequenced correctly this weekend. I digested and ligated these parts with pBjh1600. Hopefully, moving these parts from a pUC plasmid to a p15a plasmid will reduce the toxicity by lowering the copy number.
I asked Sherine and Cici to miniprep the DH10B colonies that I grew up yesterday. They also miniprepped the one BamHI colony that looked right in the colony PCR and I sent it out for sequencing today.
Since Sam's BamHI plasmid, pBca9145-I716210, was miniprepped from Righty earlier, I asked Molly to transform it into DH10B. She also transformed pBjh1600 into DH10B so that we can miniprep and replenish Jin's stock that I used today.
The transformations that I did yesterday of pBca1256 & pBjh1601KC into TG1 with pAH57 did not work well because I did the rescue at 37 degrees instead of 30 degrees. I diluted a saturated culture from yesterday and grew it to midlog & redid the transformation.
I looked at the sequencing data that I got today.
Madhvi 00:53, 29 June 2008 (UTC)
For the gateway experiment, I did the following transformations:
- pBjh1601KC into TG1 with pAH57 and pBca1256
- pAH57 into TG1 with pBca1256
- pBjh1601KC into TG1 with pBca1256
- pAH57 into TG1 with pBjh1601KC
Madhvi 23:13, 29 June 2008 (UTC)
- miniprep K112103 from DH10B
- put correct plasmids in stock plate
- colony PCR of BamHI parts
- transformations for Gateway
Madhvi 01:03, 1 July 2008 (UTC)
- pBca1256 into TG1 with pBjh1601KC and pAH57
- pBjh1601KC into TG1 with pBca1256 and pAH57
- pAH57 into TG1 with pBca1256 and pBjh1601KC
Madhvi 00:06, 2 July 2008 (UTC)
Today, my sequences for the BamHI ({r.BamHI>}, {a~r.BamHI!}, and {rbs.r.BamHI!}) came in and at least one of each clone sequenced correctly! So, I guess the trick was putting them in a p15a plasmid after all. So now I am officially done with basic parts!
Since the colony PCRs from both yesterday and Sunday did not give good results (because the thermocycler was set to do 10 repetitions instead of 30), I set up another colony PCR with the cultures that sequenced correctly in order to verify that the procedure works with the correct cycle. Indeed, the gel looked good with nice bright bands around 700bp. Since the big thermocyler (the one that is broken right now) is made by the same company, it should work just fine if we run the same cycle on it.
I am also doing a buffer screen with the new LR Clonase kit that we got. This kit has the buffer separate from the enzyme, so we can truly test the effect of the different buffers without changing the volume. The protocol that we followed is:
- Add the following to a 0.5mL tub at room temperature and mix:
- Entry Plasmid (pBjh1600): 0.33uL
- Assembly Plasmid (pBjh1601KA): 0.33uL
- 5X Buffer (LR Clonase, Gateway Buffer #1, Gateway Buffer #2,Phusion HF): 0.66uL (For Expand Buffer 3 that is 10X, add 0.33uL Buffer and 0.33uL water)
- TE Buffer, pH 8.0: 1.5uL
- Remove the LR Clonase enzyme mix from the -80C freezer and thaw on ice for about 2 minutes. Vortex the LR Clonase enzyme mix briefly twice (2 seconds each time).
- Add 0.66uL of Clonase enzyme mix to the reaction and mix well by vortexing briefly twice. Microcentrifuge briefly.
- Return LR Clonase enzyme mix to -80C storage immediately after use.
- Incubate reactions at room temperature for 1 hour.
- Add 0.33uL of Preteinase K solution to each sample to terminate reaction. Vortex briefly. Incubate samples at 37C for 10 minutes. Let DH10B competent cells thaw on ice during this incubation period.
- Add 30uL KCM and 50uL water to the competent cells. Aliquot 50uL of this mixture into each Gateway reaction mixture on ice. Remember to do a negative control (just plain DH10B cells).
- Leave on ice for 10 minutes then heat shock at 42C for 90 seconds. Put tubes back on ice for 2 minutes.
- Rescue with 100uL of 2YT and incubate in the 37C shaker for 1 hour.
- Plate of Amp/Kan plates.
The buffers that we are screening (Phusion HF, Expand 3, Gateway #1, Gateway #2) are the ones that showed apparent lysis (were viscous after 1 hour incubation with arabinose) when we did our previous experiment as well as both of the Gateway buffers we made based on publications that we found. Since this experiment called for 5x buffer, I mixed 1mL of each buffer at 5x concentration. Using our
Madhvi 04:59, 10 July 2008 (UTC)
Yesterday:
- Shirene & I recorded the screening results and did a colony PCR on the things that were picked the day before
- Since our google spreadsheet making was getting out of control, we discussed how to streamline it with Terry. We decided to fuse "digestion map analysis" and "screening & colony PCRs for transfers."
- Christie worked on updating the "screening & colony PCRs for transfers" so that we have the expected band lengths for all of the parts when we do restriction digests. She also made sure that enzymes that we use will give bands that don't overlap & give big enough bands (>300 bp).
- We made comp cells and I transformed and plated on single antibiotic plates to check whether they were okay.
- I told Molly to regrow the clone #2 cultures (from the stock plate that I streaked yesterday) that I picked yesterday for things that were cotransformed or looked wrong. (I grew a bunch of them in the wrong media yesterday, so they didn't grow)
- I told Aron to grow up colonies and streak plates (for screening and stock plates) for yesterday's transfers.
Today:
- minimeeting
- everyone miniprepped
- Dirk & I - zymo yesterday's minipreps
- digestion map with 3uL of zymo worked
- Mach 1 and DH10B comp cells that we made on Tuesday are contaminated--grew up new cultures and streaked cultures on different antibiotic strains to make sure that the strain is ok
to do:
- put results of digestion mapping online
- make tss
- put colony PCR results on google doc
- BamHI/XhoI digestion map of small set of this morning's minipreps & questionable things
Madhvi 23:00, 13 July 2008 (UTC)
Protocol for today's gateway experiment:
- Grow up cultures of Mach1 with pSB3C6-pBjh1311 to saturation
- Dilute approximately 100X (50uL culture in 5mL media) and grow to mid-log
- Spin down 600uL of culture in each 1.5mL eppendorf. In Friday's experiment, I spun down 750uL of culture in each tube
- Wash with 500uL of 1X buffer
- Estimate the volume of the cell pellet and add the appropriate amount of 5X buffer
- Add 1uL of 20% arabinose to each tube and incubate at 37C for 1 hour
- Use 0.66uL of these mixtures in place of 0.66uL 5X buffer when setting up gateway reactions (0.33uL entry vector (pBca1256), 0.33uL assembly vector (pBjh1601KC), 1.32uL water, and 0.66uL clonase enzyme)
- Pipette up and down and vortex to mix reagents
- Incubate at room temperature for 1 hour
- Terminate the reaction by adding 0.33uL of proteinase K and incubate at 37C for 10 minutes
- Transform into 50uL of DH10B competent cell mixture (combination of 220uL cells, 30uL KCm, and 50uL water)
- Rescue and plate on appropriate double antibiotic plate (since the assembly vector that I am using is KC (found in well A7 of Jin's stock plate), I plated on KC plates)
Madhvi 19:41, 16 July 2008 (UTC)
To do today:
- miniprep ig1122 and ig1096
- do transfers using these minipreps
- digestion map these minipreps
- transform entry and assembly vectors into TG1 cells with Bjh1311 (pBca1254AK and pBca1256 into TG1 with pSB3C6-pBjh1311; pBjh1600 and pBjh1601KC into TG1 with pSB1A2-pBjh1311)
- grow up colonies from assembly plates (streak on plates for screening)
- spreadsheet of assembly parts
- grow up DH10B and Mach1 for comp cells
- analyze digestion map gel from yesterday
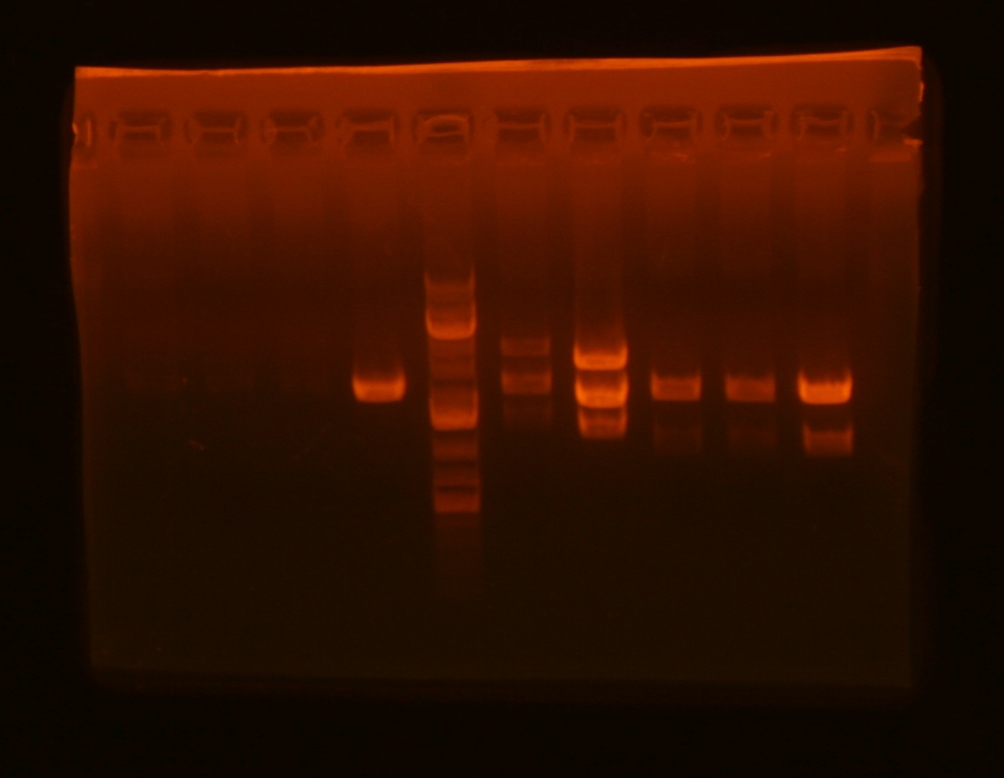
 Lane 1: ig1096 (clone#1) - BamHI/XhoI;
Lane 2: ig1122 - BamHI/XhoI;
Lane 4: pKD3 miniprep undigested;
Lane 5: pKD4 miniprep undigested
Lane 1: ig1096 (clone#1) - BamHI/XhoI;
Lane 2: ig1122 - BamHI/XhoI;
Lane 4: pKD3 miniprep undigested;
Lane 5: pKD4 miniprep undigested
Madhvi 16:01, 17 July 2008 (UTC)
to do:
- analyze gels from the past two days
- talk to Chris about finding another conditional origin of replication
- miniprep & digestion map the samples grown up yesterday
- transfer {b1006} to [A/C]
- grow up cultures from yesterday's transfers and assemblies
- grow up cultures for the other ig parts needed later in assembly
- make DH10B and Mach1 comp cells
- grow up cultures of the TG1 cells with lysis plasmid and assembly/entry plasmid for gateway expt
Madhvi 20:50, 18 July 2008 (UTC)
- analyze gels and put on wiki
- gateway expt
- Mach 1 comp cells
- miniprep & digestion map cultures from yesterday
- 3 assemblies & 2 transfers
Madhvi 22:32, 22 July 2008 (UTC)
- redo transfer of <ligase! because the sequence of mv16 showed that the part I used was actually rbs.ligase!
- miniprep, digestion map and send mv11 for sequencing
- send mv10 for sequencing
- gateway
- grow up cultures of mv4, mv7, and mv15
- cotransform pkd3 and pkd4
- test righty and lefty cells
Madhvi 20:21, 24 July 2008 (UTC)
- update sequencing log
- run gel on digestion maps of mv4, mv11, and mv15
- send out samples that digestion map correctly for sequencing
- regrow a colony for the mv1 transfer ({rbs_BglII!})
- grow up colonies of ig1094 and ig1098 from yesterday's plates
- miniprep & digestion map <ligase! cultures from yesterday
- throw away anything that has lamB and phoApp to avoid future confusion because they are wrong
- remind richard to reverse sequence mv11
- send more of MJV076 (ig114-1) for sequencing
- set up PCR for rbs.TrfA and write up construction file
Madhvi 21:34, 25 July 2008 (UTC)
- run gel for digestion map of ig1090 samples
- send one out for sequencing
- miniprep {rbs_BglII!} culture and redo mv1 transfer
- miniprep & digestion map ig1094 and ig1098 samples
- transform ig1124-1 into Righty
- email Richard about reverse sequencing for mv11
- send a different sample of pBad for sequencing
Madhvi 18:51, 26 July 2008 (UTC)
- update sequencing log
- run gel for samples of ig1094 and ig1098
- send ig1094 and ig1098 for sequencing
- pick colonies for mv1 and ig1124
- transform ig1091 into righty (plate and go directly into liquid culture)
- write construction file for rbs.TrfA & get sequence from Jin for parts spreadsheet
- email Richard about reverse sequencing for mv11--give him more DNA
tomorrow:
- analyze sequences and update sequencing log
- miniprep & digestion map mv1, ig1124, and ig1091 cultures
- 1-2-3 assembly of mv4 (through digestion and heat shock)
The sequence for ig1098 was wrong (it didn't look like it was in BBb), so I couldn't do the assembly of mv7 until yesterday. I grew up, screened, and digestion mapped a lot more clones to find one that I could use. The digestion maps looked a bit strange, but I grew them up again from the stock plate and will digestion map again and get them sequenced.
Madhvi 21:17, 29 July 2008 (UTC)
yesterday:
- 1-2-3 assembly of mv7 & do ligation and transformation for assembly of mv4
- Sent out mv1-4, ig1094-4 and ig1098-4 for sequencing; mv1-4 has been having slightly high bands on digestion maps, a couple of ig1098 clones looked right and ig1094 clones had very faint bands, but looked ok on the previous digestion map
today:
- The rbs.trfA basic part that Christie made sequenced correctly! So, I will be joining it with the b1006 terminator today.
- The oriV basic part that Chris made turned out to be some other part of the genome. New oligos will be ordered today and we should get them tomorrow.
- look at sequencing and update sequencing log
- pick colonies for mv4 and mv7
- transfer ptet into C/K
- transfer rbs_phoApp K/C; transfer rbs_lamB into C/A
- Aron found rbs_pelB yesterday, so he is transferring it into A/K; he is picking colonies today and we will miniprep and digestion map tomorrow
Madhvi 01:56, 1 August 2008 (UTC)
- miniprep and digestion map yesterday's cultures
- I need to remake the attR2 part (all of the colonies look like they are something other than E.Coli because the media used for rescue was contaminated)
- we got oligos for the OriV part, so I need to PCR that
- try to find rbs_BamHI part that is correct
- PCR rbs_BamHI for Aron
- redo assembly tree involving rbs_BamHI and do any necessary transfers
- Put the rbs_Cre and R6K parts from yesterday into appropriate vectors
Madhvi 21:05, 4 August 2008 (UTC)
- transfer rbs_BamHI
- redo R6K transfer (Pbad and rbs.Cre transfers appear to be fine)
- redo ligation and transformation for OriV (because the colonies are weird because of the Mach1 contaminant)
- redo PCR for attR2
- pick colonies for double antibiotic parts
- pick more colonies for mv15 and mv10
- send out mv23-1 and mv16-1 for sequencing
- miniprep mv10-2 and send for sequencing tomorrow(the other two clones of mv10 and all three clones for mv15 were cotransformed)
- restreak pCP20 on AC plate from -80 stocks (and go directly into culture)--grow at 30C because of temp sensitive origin
Madhvi 03:27, 9 August 2008 (UTC)
- design oligo for attR2
- redo OriV PCR
- grow new cultures of comp cells and throw away the old ones
- grow up pG80ko-mv23 culture (2 colonies)
- miniprep & digestion map (mv4, mv12, and mv21)
- PCR rbs.BamHI to make sure it is in the right assembly vector (I am not sure whether it is in KA or AK)
- grow up mv15
tomorrow:
- assemblies of mv5, mv13, and mv22
- miniprep and digestion map mv15
- DBBS for mv17 (PCR mv15 and mv16)
- miniprep, digestion map & transform pG80ko-mv23 plasmid
- digest, ligate transform, OriV
- send double antibiotic parts for sequencing of attR2 site
Madhvi 08:18, 16 August 2008 (UTC)
PCRs set up:
- diagnostic for TrfA strain:
- ca604F/ca604R on MC1061::mv23 (4K55)
- JH102F/JH102R on MC1061::mv23 (55)
- OriV version of entry vector:
- mv041F/mv038 on pBca1256-OriV (55)
- mv034/ca1092R on pBca1256 (4K55)
Madhvi 02:09, 20 August 2008 (UTC)
- redo K112239
- ligate & transform DBBS (mv6 & mv14)
- redo mv22
- Dilute OriV entry vector 25 times and test it by transforming into TrfA strain and TG1. I mixed the dilution with molly 19 diluted 25 times and transformed 1uL of the resulting mixture into both strains.
Madhvi 04:28, 22 August 2008 (UTC)
The OriV entry vector appears to be wrong because it also survived in TG1 (there were tons of red colonies on that plate). In fact, there were more red colonies on the TG1 plate than the TrfA strain plate. I digestion mapped the OriV entry vector alongside the OriV entry vector including the pUC origin to make sure that it was indeed removed (I suspect that the pUC origin is still there). I also ran mapped pBca1256 alongside the two to confirm that the OriV part was actually incorporated. I used BamHI/XhoI for all of the digests and expected the following bands: 1142+2040 for the OriV vector without the pUC origin, 1896+2040 for the OriV vector with the pUC origin and 1486+1896 for pBca1256.
There were no colonies on the mv6 and mv14 plates that were constructed using dbbs. Molly and Aron have been having trouble with their dbbs assemblies as well, so it seems that there is something systematically wrong. The mv22 plate has a few colonies which I will screen using colony PCR with ca1091F (forward ptet oligo) and g00101. The K112239 plate has many colonies. I will screen a number of them using ca998 and dr016 (reverse attR2 oligo). Since I won't be in tomorrow, Molly will miniprep the correct cultures tomorrow and can move to the next step of assembly for the parts.
Madhvi 21:07, 26 August 2008 (UTC)
- things to transfer:
- mv5-1, mv13-1, mv21, mv16, mv15-1, mv15-2 into pBca1256
- rbs_ligase (K112109) into pBjh1600
- sequence clones 1 and 2 of mv15
- sequence more clones of K112120 [A/K]
- tranform pBca1256-Bca1091 (ptet) into lefty to amplify
- assemble R6K and attR2 rev
- make promoter from pAH57
- PCR, digest, ligate to remove pUC origin from OriV vector
Madhvi 16:25, 27 August 2008 (UTC)
- transform R6K and attR2 rev into Righty/Lefty so that I can repeat assembly with background subtraction
- transform pBjh1600-K112120 into Righty to amplify
- tranform mv16-1 into Lefty so that it can be transferred into pBjh1600 (mv15 can be transferred into pBjh1600 using PCR once it is confirmed)
- transform pBca1256-Bca1109 (Pbad) into lefty from Chris's working plate so that it can be used for assemblies and dbbs
- transfer K112109 into pBjh1600 and transform into lefty for assembly with pBjh1600-K112120
- pick colonies from yesterday's plates (OriV vector, mv5, mv13, mv21, mv27, promoter)
- genomic prep and PCR with 603F/R of MC1061 with pInt to check if the two extra bands we saw last time are just things from that genome
Madhvi 22:39, 30 August 2008 (UTC)
Yesterday, I did a bunch of assemblies (mv15, mv27, mv28, mv30) and transferred Bca1109. All of these plates have colonies on them, but mv15 only has a few. Since the pAH57 promoter (K112126) sequenced correctly, we are fine with the assembly that we did. Also, mv27 (R6K.attR2 rev) sequenced fine, so the assembly of mv28 should be fine. Yesterday, I also redid the OriV vector digesting with DpnI to remove the background from the parent vector containing a pUC origin. It didn't really work and Chris suspects that the strain is wrong. There are four things that could be wrong: either the strain or the plasmid are not the things we actually intended to make or they are what we intended but aren't functional. From the weird 603F/R PCR result, Chris is sure that the strain is not what we intended to make and suspects that it is not functional. He thinks that the apparent presence of TrfA from PCR with those oligos might be a false positive. The MC1061-pInt competent cells that I used are probably bad (Sam was having trouble with her genomic integrations as well). Chris wants me to remake the strain starting from MC1061 and putting in pInt. He also wants me to test the current strain by making a functional OriV plasmid from the Fosmid lucigen kit that we ordered.
Here's what I am doing today:
- miniprep and digestion map the transfers of mv13 and K112109 (these were transferred into pBca1256 and pBjh1600, respectively)
- Assemble K112126 (pAH57 promoter) with pBca1256-mv21
- Make OriV vector from Fosmid kit (transform using epicenter cells)
- Assemble pBjh1600-K112109 (rbs.ligase) with pBjh1600-K112120 (rbs.BamHI)
- pick colonies for mv15 (rbs_phoApp.BamHI), mv28 (KA part.R6K.attR2 rev), mv30 (Phagemid.attR1.R6K.pAH57 promoter), Bca1109 (Pbad)
- grow up a culture of MC1061 with pInt from Sam's plate (in case she doesn't get back to me tomorrow about where her comp cells are)
The K112109 and K112120 parts were single cut when I ran my gel, so I'll try that assembly again tomorrow. The bands for K112126 and mv21 were weird when I ran my gel (it looks like I might have switched these two samples), but I went ahead and used the large band for each of these. The large band on all of the mv13 samples also looked low.
| Lane | plasmid | clone | enzyme 1 | enzyme 2 | expected bands | result |
| 1 | pBjh1600-K112109 | 1 | BamHI | AlwNI | 4413 + 413 | single cut |
| 2 | pBjh1600-K112109 | 2 | BamHI | AlwNI | 4413 + 413 | single cut |
| 3 | pBjh1600-K112109 | 3 | BamHI | AlwNI | 4413 + 413 | single cut |
| 4 | pBjh1600-K112120 | BglII | AlwnI | 2924 + 1080 | single cut | |
| 5 | pBca1256-K112126 | BamHI | BglI | 2048 + 1316 | bands look like mv21 | |
| 6 | pBca1256-mv21 | 1 | BglII | BglI | 3072 + 1994 | bands look like K112126 |
| 7 | pBca1256-mv13 | 1 | EcoRI | XhoI | 4149 + 1133 | large band too small |
| 8 | pBca1256-mv13 | 2 | EcoRI | XhoI | 4149 + 1133 | large band too small |
| 9 | pBca1256-mv13 | 3 | EcoRI | XhoI | 4149 + 1133 | large band too small |
Protocol for construction of Fosmid OriV vector:
- Digest on 100uL scale (use 10uL NEB2, 84uL water, 5uL MG1655 genomic prep, 1uL EcoRV). Incubate at 37C for 1 hour.
- Zymo cleanup by adding 300uL of ADB buffer and doing two washes with 200uL each. Elute with 6uL of water.
- Ligate with 1uL EcoRV digest, 1uL Lucigen Fosmid blunt vector, 1uL Ligation Buffer, 6.5uL water, and 0.5uL T4 DNA ligase. Incubate at room temperature for 30 minutes.
- Add one aliquot of epicentre TrfA cells to ligation and leave on ice for 30 minutes.
- Heat shock for 30 seconds at 42C.
- Incubate 2 minutes on ice and then rescue with 100uL 2YT and 0.2uL of inducer 1000X.
- Plate on Cm plate.
To finish testing the plasmid that I make through this process and to test my strain, I will:
- grow 1 colony and split it in two; add inducer to only one
- miniprep cultures and run gel; hopefully, only the culture which was grown at the inducer will show a band and the other will be empty
- transform into my trfA cells and grow colony
- miniprep & run gel to show that it is high copy
Madhvi 00:24, 1 September 2008 (UTC)
- retry the assembly of K112109 and K112120
- redo assembly of K112126 and mv21--remember to rescue and plate at 30C
- digestion map mv28, Bca1109 (pBad), and mv15 (and mv13)
- dbbs assembly of Bca1109 with mv5 and mv13
- assemble mv30 with K112239 (rbs_xis.int.ihfA.ihfB.attR2) and K112240 (rbs_xis.int.attR2)--remember to rescue and incubate at 30C
Madhvi 03:38, 25 September 2008 (UTC)
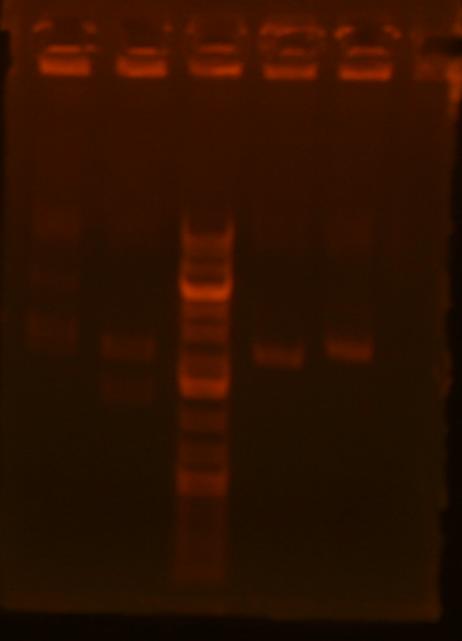
So, I digestion mapped and mv39-clone#1 looks right. I will confirm by sequencing tomorrow. Also, mv23-Bjh1233 clones #2-4 looks like they might be right, but I can't see the small band. mv37: all clones look wrong. Here's the gel pic:
| Lane | plasmid | clone | enzyme 1 | enzyme 2 | expected bands | result |
| 1 | pBca1256-mv39 | 1 | XbaI | HindIII | 6261+620 | right! |
| 2 | pBca1256-mv39 | 2 | XbaI | HindIII | 6261+620 | wrong |
| 3 | pBca1256-mv39 | 3 | XbaI | HindIII | 6261+620 | wrong |
| 4 | pBca1256-mv39 | 4 | XbaI | HindIII | 6261+620 | wrong |
| 5 | pBca1256-mv39 | 5 | XbaI | HindIII | 6261+620 | wrong |
| 6 | pBca1256-mv39 | 6 | XbaI | HindIII | 6261+620 | wrong |
| 7 | pBca1256-mv39 | 7 | XbaI | HindIII | 6261+620 | wrong |
| 8 | pBca1256-mv39 | 8 | XbaI | HindIII | 6261+620 | wrong |
| 9 | pBca1256-mv37 | 1 | NcoI | XbaI | 4261+441 | wrong |
| 10 | pBca1256-mv37 | 2 | NcoI | XbaI | 4261+441 | wrong |
| 11 | pBca1256-mv37 | 4 | NcoI | XbaI | 4261+441 | wrong |
| 12 | mv23-Bjh1233 | 1 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 13 | mv23-Bjh1233 | 2 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 14 | mv23-Bjh1233 | 3 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 15 | mv23-Bjh1233 | 4 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
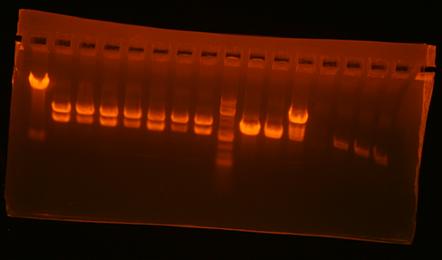
Madhvi 20:17, 25 September 2008 (UTC)
| Lane | plasmid | clone | enzyme 1 | enzyme 2 | expected bands | result |
| 1 | pBca1256-mv39 | 1 | BamHI | XhoI | 5395+1486 | right! |
| 2 | pBca1256-mv37 | 1 | BamHI | XhoI | 3216+1496 | wrong |
| 3 | pBca1256-mv37 | 2 | BamHI | XhoI | 3216+1496 | wrong |
| 4 | pBca1256-mv37 | 4 | BamHI | XhoI | 3216+1496 | wrong |
| 5 | pBca1256-Bca1172 | BglII | XhoI | 904+2057 | ||
| 6 | pBca1256-mv23 | XbaI | HindIII | 6261+620 | wrong | |
| 7 | pBca1256-mv39 | 7 | XbaI | HindIII | 6261+620 | wrong |
| 8 | pBca1256-mv39 | 8 | XbaI | HindIII | 6261+620 | wrong |
| 9 | pBca1256-mv37 | 1 | NcoI | XbaI | 4261+441 | wrong |
| 10 | pBca1256-mv37 | 2 | NcoI | XbaI | 4261+441 | wrong |
| 11 | pBca1256-mv37 | 4 | NcoI | XbaI | 4261+441 | wrong |
| 12 | mv23-Bjh1233 | 1 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 13 | mv23-Bjh1233 | 2 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 14 | mv23-Bjh1233 | 3 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
| 15 | mv23-Bjh1233 | 4 | EcoRI | BamHI | 506+2107 | possibly right (can't see small band) |
 "
"