User:Jec105
From 2008.igem.org
m (New page: {{Imperial/StartPage}} __NOTOC__ Bad name, I know... But half the fun of iGEM is poor punning! <hr> <center>Welcome to the Imperial 2008 i...) |
m |
||
| Line 1: | Line 1: | ||
| - | {{Imperial/ | + | {{Imperial/StartPage2}} |
| - | + | === Summer Summary === | |
| + | {{Imperial/Box2|| | ||
| + | This page will include... | ||
| - | + | Genetic diagrams, engineering cycle, overview of modelling, overview of major results, contributions etc. | |
| + | Basically just an overview of our project progress and results, with a section detailing how we achieved a Gold medal!|}} | ||
| - | + | {{Imperial/Box1|Our Approach| | |
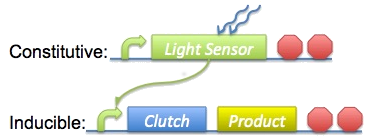
| - | + | [[Image:Imperial_2008_Basic_Circuit.png|300px|right]] | |
| - | + | Our basic approach was to constitutively upregulate a light-sensitive pathway in ''B. subtilis'', then use a downstream product from that pathway to selectively regulate expression of our clutch and biomaterial. This overview is shown on the right. To do this we need to produce at least one constitutive promoter to precede our light-sensing mechanism, as well as a 'light-inducible' promoter in front of our clutch and biomaterial. Additionally, we'll need to make all the other standard parts such as ribosome binding sites (RBS) and terminators. | |
| - | + | ||
| - | -- | + | |
| - | + | ||
| + | The diagram below shows our theoretical final construct once it's all put together - as you can see, it's a little more complex than the one on the right! | ||
| - | + | <html><center><img width=750px src="http://s59.photobucket.com/albums/g305/Timpski/S1L.png"></center></html> | |
| - | + | ||
| - | + | Here, AB is our antibiotic resistance cassette, ''ytvA'' is the gene controlling the light-sensing pathway, ''SB'' is the biomaterial, ''epsE'' the clutch and the 5' and 3' sections are integration sites. Light-inducible promoters are labelled with an 'L'. | |
| - | + | |}} | |
| - | + | ||
| - | | | + | {{Imperial/Box1|Dry Lab Overview| |
| - | | | + | Basically our dry lab team concentrated on characterising the chassis. In the dry lab section you'll find pages on the genetic circuit, growth curve and motility analysis from our project; this section will give a brief brief overview of each area (no pictures?). It will also include (in the motility analysis part) a movie (y/n?) of the motility response of ''B. subtilis'' with an inducible ''epsE'' gene BioBricked in. |
| - | + | ====== Growth Curve ====== | |
| - | + | The growth curve of ''B. subtilis'' was modelled by superposition of three more basic ODE models, which were constructed and simulated in MATLAB. The lag, exponential and stationary phases are modelled and combined to produce the curve on the right (Image here?). | |
| - | + | ====== Genetic Circuit ====== | |
| - | + | We feel accurate modelling of the genetic circuit contributes greatly to the characterisation of synthetic systems. As part of the project, the behaviours of constitutive and inducible promoters were modelled for comparison with our experimental data. | |
| - | + | ====== Motility Analysis ====== | |
| - | + | A major component of the system is the motility and shift between a motile and arrested state with the expression of EpsE. | |
| - | | | + | |}} |
| - | + | ||
| - | <br | + | {{Imperial/Box1|Wet Lab Overview| |
| - | [[Team:Imperial_College/ | + | The wet lab team was responsible for designing the constructs and setting up the cloning strategy to get us from the starting parts to the finished system. We were also responsible for designing and BioBricking the starting parts, of course, and designing and implementing the integration brick technique. This section shows some of the major results that came from the wet lab over the summer. Motility results can go in the dry lab overview above. |
| - | {{Imperial/EndPage}} | + | ====== Transformation ====== |
| + | Transformation stuff goes here... | ||
| + | ====== Calibration Curve ====== | ||
| + | Calibration curve results go here... | ||
| + | |Stuff}} | ||
| + | |||
| + | {{Imperial/Box1|Achievements| | ||
| + | *Helped Bristol by sending them a mini-iGEM project: ''Chemotactic dot-to-dot'' with information on quorum sensing and directed movement | ||
| + | *Helped Bristol by sending them a part (BBa_J37015) from our 2007 stock which was an empty vector in the Registry | ||
| + | *Developed integration bricks, to allow devices to be constructed that can then be excised and planted into ''B. subtilis'' | ||
| + | *Layed the groundwork for future teams to work with ''B. subtilis'' by BioBricking promoters, RBSs, terminators and so on and characterising them | ||
| + | *Showed that expansion into other organisms is a definite possibility! | ||
| + | |<html><align="right"><img width="100px" src="http://i59.photobucket.com/albums/g305/Timpski/Gold_Medal.png"></align></html>}} | ||
| + | |||
| + | <hr><br> | ||
| + | {{Imperial/Box2||Of course, that's a very simplified description of our project. We expanded upon our project by looking into possible areas for real-world application; for a case-study of such an implementation check out how our project fits in with [[Team:Imperial_College/Biocouture | '''>>> Biocouture >>>''']]}} | ||
| + | {{Imperial/EndPage|Chassis_2|Biocouture}} | ||
Revision as of 20:44, 20 October 2008
Summer Summary
|
|||||||||||||||||||||||||||
 "
"