Team:Michigan/Project/Modeling
From 2008.igem.org
(→Sequestillator Modeling) |
|||
| (28 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<font color=navy> | <font color=navy> | ||
| - | = '''<font color= | + | = '''<font color=royalblue size=6>Sequestillator Modeling</font>''' = |
| - | The | + | The creation of a synthetic genetic clock has been considered by many people to be a "holy grail" of synthetic biology. Elowitz's "Repressilator" was one of the first synthetic clocks constructed, and it paved the way for consideration of other topologies that may promote oscillatory behavior, such as the Atkinson clock. |
| - | + | A common characteristic of genetic clocks in general is the presence of a negative feedback loop (i.e. A activates B, and B represses A, so by repressing A, B indirectly its own concentration). In the Repressilator and the Atkinson clock, this repression occurred on the pre-transcriptional level; that is, the repressor protein prevented transcription of the activator gene. | |
| - | + | Below, we present three topologies of clocks with their modes of repression. | |
| - | + | ||
| - | + | ||
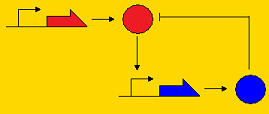
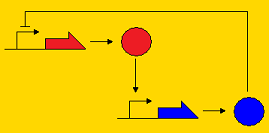
<div align='center'>[[Image:Picture8.png]] [[Image:Picture3.png]] [[Image:Picture2.png]]</div> | <div align='center'>[[Image:Picture8.png]] [[Image:Picture3.png]] [[Image:Picture2.png]]</div> | ||
<div align='center'>Post-translational repression Pre-translational repression Pre-transcriptional repression </div> | <div align='center'>Post-translational repression Pre-translational repression Pre-transcriptional repression </div> | ||
| - | + | ||
| + | Note that the pre-transcriptional repression topology is the Atkinson clock without the positive feedback loop, and the post-translational clock is the Sequestillator. | ||
| + | One of the goals of the modeling team is to understand the dynamics of this repression loop - i.e., how does the type of repression effect the robustness of the oscillator and the behavior (amplitude, periodicity, etc.). | ||
| + | |||
| + | Analytically, it's pretty tough to answer these questions due to the multiple nonlinearities associated with the ODEs. However, numeric techniques can prove useful in helping answer our questions about robustness and behavior. We can pick lots of different, physiologically relevant parameter sets, put them into our ODEs, and see what trajectories we get. Particularly, going back to our question about robustness, we can assign a value for robustness. We define this value as the total number of parameter sets that give oscillatory behavior over all possible feasible parameter sets. Within our lab, we call this value the Ninfa index. | ||
| + | |||
| + | |||
| + | <font size=4 color=royalblue>Our general algorithm for "selecting" for oscillations is as follows:</font> | ||
| + | *For a parameter set, use ODE Solver to get trajectories. Take one of those trajectories, select a time period that allows the dynamics to evolve appropriately (i.e. chose a time period that is several generations away from t=0) and do the following: | ||
| + | * Apply Fast Fourier Transform on trajectory to get a Power Spectral Density - there should be two symmetric peaks if there is a chance for oscillatory behavior. However, sometimes damped oscillations creep and the PSD shows two peaks. | ||
| + | * Low Pass filter: max sure the maximum Fourier coefficient is larger than a specific, relatively small value. This gets rid of really, really small oscillations that may be attributable to numeric errors, and wouldn't be detected experimentally anyways. | ||
| + | *Damping Test: detects amplitude of oscillations and removes them if the amplitude decreases and time increases (these would be damped oscillations) | ||
| + | |||
| + | |||
| + | We created the "Indexilator" to calculate "relative" Ninfa indices (instead of dividing by the total feasible range of parameter space, divide by the number of trials you ran). | ||
| + | The "Indexilator" is coded in Mathematica (we are currently working on programming a MatLab version). A commented copy of the code, followed by an example trial are given below. Anyone should free to utilize this code. | ||
| + | |||
| + | *[https://2008.igem.org/wiki/index.php/Image:indexilator.nb <font color=blue>Indexilator in PDF format</font>] <br> | ||
| + | *For some reason, I can't put up Mathematica files on this wiki, so go [http://synthbio.engin.umich.edu/wiki/index.php/Image:Indexilator.nb here] to get the code. | ||
| + | *[https://2008.igem.org/wiki/index.php/Image:Example.pdf <font color=blue>Sample ODE in Indexilator:PDF format</font>] <br> | ||
| + | *[http://synthbio.engin.umich.edu/wiki/index.php/Image:Example.nb Sample Indexilator code]: Example does randomized searches of different parameters | ||
| + | |||
| + | |||
| + | Two Quick Notes on Randomized Searches: | ||
| + | * Mathematica's random number generator is very good - If you look for a random number between a and b, each number in between a and b has pretty much an equal probability of showing up. So, if you're search between 0 and 15, there's a 1/3 probability your values will be between 0 and 5, and 2/3 probability they will be between 5 and 15. | ||
| + | * Most of the indices on the wiki are the result of randomized searches. | ||
| + | |||
| + | |||
| + | ==<font size=5 color=royalblue>Sequestillator Modeling Pages</font>== | ||
| + | |||
| + | These pages reference the different models that have been considered: | ||
| + | *[[Team:Michigan/Project/Modeling/Model1 | Model 1]] | ||
| + | *[[Team:Michigan/Project/Modeling/Model2 | Model 2]] | ||
| + | *[[Team:Michigan/Project/Modeling/Model3 | Model 3]] | ||
| + | |||
| + | |||
| + | |||
| + | ==<font size=5 color=royalblue>Other Simulations That Will Appear at iGEM 2008 (in less than two weeks!)</font>== | ||
| + | |||
| + | I couldn't include a lot of other simulations on the wiki due to my lack of wiki-editing skills - but have no fear, for at iGEM they shall appear! We will have more Ninfa Index calculations and cloud pictures(and other simulations!) on our poster, so be sure to stop by to see our modeling! | ||
| + | |||
| + | *Ninfa Index comparisons (and cloud pictures!) of the three oscillators described here | ||
| + | *Ninfa Index comparisons (and cloud pictures!) of modified versions of the three oscillators (add in positive feedback loop) | ||
| + | *Stochastic comparisons of all three oscillators | ||
| + | *Comparison of the robustness of the actual mammalian clock topology to ours | ||
| + | [https://2008.igem.org/Team:Michigan/Project Back to Project Home] | ||
|} | |} | ||
Latest revision as of 02:52, 30 October 2008
|
|---|
|
Sequestillator ModelingThe creation of a synthetic genetic clock has been considered by many people to be a "holy grail" of synthetic biology. Elowitz's "Repressilator" was one of the first synthetic clocks constructed, and it paved the way for consideration of other topologies that may promote oscillatory behavior, such as the Atkinson clock. A common characteristic of genetic clocks in general is the presence of a negative feedback loop (i.e. A activates B, and B represses A, so by repressing A, B indirectly its own concentration). In the Repressilator and the Atkinson clock, this repression occurred on the pre-transcriptional level; that is, the repressor protein prevented transcription of the activator gene. Below, we present three topologies of clocks with their modes of repression. Post-translational repression Pre-translational repression Pre-transcriptional repression
Analytically, it's pretty tough to answer these questions due to the multiple nonlinearities associated with the ODEs. However, numeric techniques can prove useful in helping answer our questions about robustness and behavior. We can pick lots of different, physiologically relevant parameter sets, put them into our ODEs, and see what trajectories we get. Particularly, going back to our question about robustness, we can assign a value for robustness. We define this value as the total number of parameter sets that give oscillatory behavior over all possible feasible parameter sets. Within our lab, we call this value the Ninfa index.
Sequestillator Modeling PagesThese pages reference the different models that have been considered:
Other Simulations That Will Appear at iGEM 2008 (in less than two weeks!)I couldn't include a lot of other simulations on the wiki due to my lack of wiki-editing skills - but have no fear, for at iGEM they shall appear! We will have more Ninfa Index calculations and cloud pictures(and other simulations!) on our poster, so be sure to stop by to see our modeling!
|
|---|
 "
"