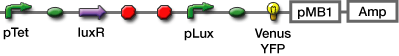Team:Chiba/Experiments:Reporter
From 2008.igem.org
(→Method) |
(→Reporter) |
||
| Line 13: | Line 13: | ||
==Reporter== | ==Reporter== | ||
| + | Our delayed switch has to turn on or off without variable time lag | ||
| + | after the preset time has passed. Therefore we need the output module, | ||
| + | which announces quickly that a particular concentration of the | ||
| + | signaling molcules has been reached. | ||
| + | We imagined three kinds of output: | ||
| - | |||
| - | + | * Fluorescent Protein | |
| + | * Bioluminescence(luciferase) | ||
| + | * Cell staining (LacZ and X-gal assay) | ||
| - | |||
| - | |||
| - | |||
| + | and tested Fluorescent Protein and β-gal(lacZ). | ||
| - | We | + | We selected GFP, Venus YFP, and mCherry. The latter two were chosen |
| + | because Venus YFP matures quickly | ||
| + | [http://www.nature.com/nbt/journal/v20/n1/full/nbt0102-87.html|(1)], | ||
| + | and we can easibly recognize mCherry coloration | ||
| - | + | [http://www.ncbi.nlm.nih.gov/pubmed/15558047|(2)]. | |
| - | + | ||
| + | |||
| + | We used fluorescent proteins and β-gal(lacZ) as a reporter. | ||
| + | |||
| + | |||
| + | |||
| + | We were able to recognize chemical luminescence of luciferase only at | ||
| + | dark place devoid of light. Therefore the conditions of use are | ||
| + | limited, and we did not select Luciferase as our reporter. | ||
==Experiment== | ==Experiment== | ||
Revision as of 03:42, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Reporter
Our delayed switch has to turn on or off without variable time lag after the preset time has passed. Therefore we need the output module, which announces quickly that a particular concentration of the signaling molcules has been reached.
We imagined three kinds of output:
* Fluorescent Protein * Bioluminescence(luciferase) * Cell staining (LacZ and X-gal assay)
and tested Fluorescent Protein and β-gal(lacZ).
We selected GFP, Venus YFP, and mCherry. The latter two were chosen because Venus YFP matures quickly
[http://www.nature.com/nbt/journal/v20/n1/full/nbt0102-87.html|(1)],
and we can easibly recognize mCherry coloration
[http://www.ncbi.nlm.nih.gov/pubmed/15558047|(2)].
We used fluorescent proteins and β-gal(lacZ) as a reporter.
We were able to recognize chemical luminescence of luciferase only at dark place devoid of light. Therefore the conditions of use are limited, and we did not select Luciferase as our reporter.
Experiment
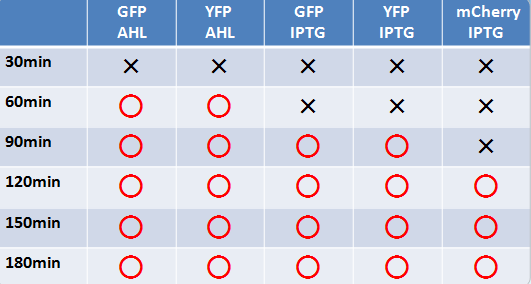
Measure the time course form adding inducer (IPTG or AHL) to confirm the change of color to analysis the gene expression.
Evaluate following gene:
- Fluorescent Protein
- GFP
- Venus YFP
- mCherry
- β-gal (X-gal assay)
- Fluorescent Protein
- GFP
- Venus YFP
- mCherry
- β-gal (X-gal assay)
Result & Discussion
Result LacZ
- 蛍光たんぱくではないLacZは目視のみで発現を確認した
- LacZはX-galの濃度によって目視での確認時間が変わった
- 液体で4時間以内に目視で確認することはできなかった
- -->8時間後目視で確認できた
蛍光たんぱく
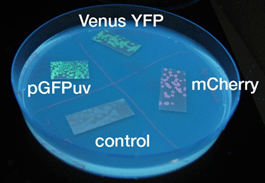
- 液体、固体培地において似たような結果になった
- 目視で一番見やさはmCherry,GFP,YFPの順であった
- 固体培地のほうが目視で確認しやすい
- AHLでのインダクションのほうがIPTGより確認できた時間が早かった
- GFP,YFPのほうがmCherryより確認できた時間が早かった
Discussion
このプロジェクトでは、発現するまでの時間を変えることが目的である
- X-galの濃度を高くすると発現を確認できるまでの時間は短くなったが、蛍光たんぱくで発現を確認できるまでの時間より遅い
- X-galを基質とするβ-galでは、時間がたつごとに基質濃度が下がり、出力までの時間が長くなってしまうことも考えられる
以上の2つの理由から蛍光タンパク質が適している
またGFP,YFPの発現時間までのタイムラグが一番少なかった
- 目視で確認した際、GFPのほうがYFPよりも目視で確認するのが容易であった
以上の理由から、出力はGFPとする。
Method
Strain:XL10G KanR
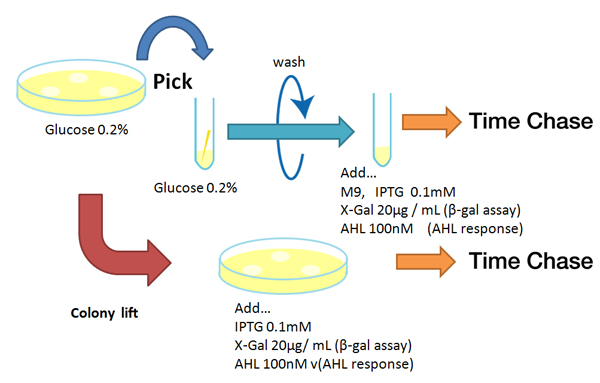
- Agar plate experiment
- Pre-culture
- Picked and cultured the following plate stocks in 2mL of LB:
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Picked and cultured the following plate stocks in 2mL of LB:
- Spread on plate
- Spread on new plate
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Spread on new plate
- Colony lift
- Colony lift to inducible agar plate (containing IPTG or AHL)
- LB-Amp+0.2 mM IPTG agar plate, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp+100 nM AHL agar plate, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Incubate at 37 °C
- Colony lift to inducible agar plate (containing IPTG or AHL)
- Check expression every 30 min.
- Liquid medium experiment
- Pre-culture
- Picked and cultured the following plate in 2mL of LB:
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Picked and cultured the following plate in 2mL of LB:
- Culture
- Dilute pre-cultures and add to new LB medium.
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for about 6 h
- Dilute pre-cultures and add to new LB medium.
- Wash
- Transfer 10mL each of the culture to 50mL centrifuge tubes.
- Centrifuged for 6min at 3600rpm,20°C and discarded the supernatant.
- Add physiological saline and resuspention
- Repeated wash twice.
- Add M9 minimal medium.
- Mix
- Dispens each culture into 48-well deep well or 96-well deep well
- Add IPTG fainal conc. 0.2 mM (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- Add AHL fainal conc. 100nM ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Measure fluorescence intensity every 1 h.
- Equipment
- shaking incubator
- Innova 4200 Benchtop or Floor-Stackable Incubator Shaker(37°C)
- 46-well plate(deep well)
- 96-well plate(deep well)
- Fluoroskan Ascent 2.5(program:Ascent Software Version 2.6)
- Beckman Allegratm X-12R Centrifuga(Beckman Coulter)
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"