Team:Michigan/Project/LandingPads
From 2008.igem.org
(New page: __NOTOC__ {{Michigan Header}} {|class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 1px; background-color:dodgerblue; border: 1px solid mediumblue; text-align:...) |
|||
| (5 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{|class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 1px; background-color:dodgerblue; border: 1px solid mediumblue; text-align:center" | {|class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 1px; background-color:dodgerblue; border: 1px solid mediumblue; text-align:center" | ||
| - | !width="10%" align=" | + | !width="10%" align="justify" valign="top" style="background:gold; color:black"| |
<font color=navy> | <font color=navy> | ||
| - | = '''<font color= | + | = '''<font color=royalblue size=6>BioBrick Landing Pads</font>''' = |
| - | ==<font size= | + | <font size=3>We will be using landing pads to insert our clock onto the chromosome of <i>E. coli</i>. Noisy behavior has proven detrimental to clock studies throughout the past, and we hope to reduce the noise in our system using landing pads. </font> |
| + | |||
| + | |||
| + | ==<font size=4 color=royalblue>BioBrick Landing Pad Polylinker</font>== | ||
{| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color:transparent; border: 1px solid transparent;text-align:center" | {| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color:transparent; border: 1px solid transparent;text-align:center" | ||
| - | !width="70%" align=" | + | !width="70%" align="justify" valign="top" style="background:transparent; color:navy"| |
| + | |||
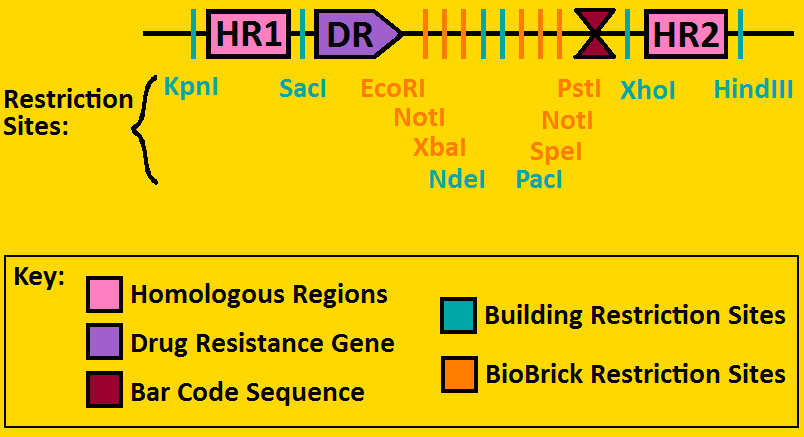
| + | <font color=royalblue>Landing Pads:</font> aid in the insertion of biological constructs onto the chromosome of <i>E. coli</i> | ||
| + | |||
| + | <font color=royalblue>Landing Pad Goals:</font> | ||
| + | * BioBrick compatibility | ||
| + | * Easy phenotypic screening | ||
| + | * Easy sequencing | ||
| + | * Limit noise | ||
| - | + | <font color=royalblue>Polylinker Features:</font> | |
| + | * <font color=orangered>BioBrick compatible restriction sites</font> - these allow for standard BioBrick construction in forward and reverse directions | ||
| + | * <font color=lightseagreen>Additional building restriction sites</font> - these are for building the landing pad, as well as changing the homologous regions and drug resistance gene (can also be used for building if needed) | ||
| + | * <font color=firebrick>Bar code sequence</font> - a 20 bp segment that allows for easy sequencing | ||
| + | * <font color=blueviolet>Drug resistance gene</font> - this allows for easy phenotypic screening - our landing pads use Chloramphenicol resistance genes | ||
| + | * <font color=deeppink>Homologous regions</font> - these specify where on the chromosome you wish to put your constructs. | ||
!width="10%" align="left" valign="center" style="background:#transparent; color:black"| | !width="10%" align="left" valign="center" style="background:#transparent; color:black"| | ||
| Line 23: | Line 39: | ||
|} | |} | ||
| - | ==<font size= | + | |
| + | |||
| + | ==<font size=4 color=royalblue>Landing Pads used for the Sequestilator</font>== | ||
| + | |||
| + | |||
| + | {|class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 1px; background-color:dodgerblue; border: 1px solid mediumblue; text-align:center" | ||
| + | !width="10%" align="justify" valign="top" style="background:gold; color:navy"| | ||
| + | |||
| + | ==<font color=royalblue size=3>Operon</font>== | ||
| + | <br><br><font size=3>Activator Operon</font><br><br><br> | ||
| + | |||
| + | == == | ||
| + | <br><br><font size=3>Repressor Operon</font> | ||
| + | |||
| + | !width="10%" align="justify" valign="top" style="background:gold; color:navy"| | ||
| + | |||
| + | ==<font color=royalblue size=3>Topology</font>== | ||
| + | |||
| + | <div align=center>[[Image:New 1st operon - gold.PNG|220px]]<br><br> | ||
| + | |||
| + | == == | ||
| + | <br>[[Image:New 2nd operon - gold.PNG|300px]]</div> | ||
| + | |||
| + | !width="10%" align="justify" valign="top" style="background:gold; color:navy"| | ||
| + | |||
| + | ==<font color=royalblue size=3>Landing Pad</font>== | ||
| + | |||
| + | <div align=left><font size=3>Leucine Landing Pad</font></div><br>constructed by former Ninfa lab member, Don Eun Chang | ||
| + | <br> | ||
| + | |||
| + | == == | ||
| + | <div align=left><font size=3>Arabinose Landing Pad</font></div><br> iGEM 2007 project by Alyssa Delke & Khalid Miri | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
{| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color:transparent; border: 1px solid transparent;text-align:center" | {| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color:transparent; border: 1px solid transparent;text-align:center" | ||
| - | !width="70%" align=" | + | !width="70%" align="justify" valign="center" style="background:transparent; color:black"| |
| - | + | ==<font color=royalblue size=3>2nd Operon in Arabinose Landing Pad</font>== | |
| + | |||
| + | This is what our repressor operon would look like in our Arabinose BioBrick landing pad. | ||
| + | |||
| + | |||
| + | Notice that the landing pad plasmids are Ampicilin resistant, to allow for additional screeining. There is also a ScaI restriction site located in the AmpR gene, that is found nowhere else on the plasmid, that can be used for linearizing the plasmid. It is necessary to linearize the plasmid in order to cross it onto the chromosome. | ||
| + | |||
| + | |||
| + | For testing and results on these landing pads, please visit last year's iGEM page on Landing Pads [http://synthbio.engin.umich.edu/wiki/index.php/BioBrick_Landing_Pad HERE]. | ||
!width="10%" align="left" valign="center" style="background:#transparent; color:black"| | !width="10%" align="left" valign="center" style="background:#transparent; color:black"| | ||
| - | <div align=center> [[Image:Landing pad plasmid - gold.png| | + | <div align=center> [[Image:Landing pad plasmid - gold.png|550 px]] |
</div> | </div> | ||
| Line 42: | Line 101: | ||
<font color=navy size=3>Learn more about Landing Pads [http://synthbio.engin.umich.edu/wiki/index.php/BioBrick_Landing_Pad <font size=3> Here!</font>] | <font color=navy size=3>Learn more about Landing Pads [http://synthbio.engin.umich.edu/wiki/index.php/BioBrick_Landing_Pad <font size=3> Here!</font>] | ||
</font></div> | </font></div> | ||
| + | <br><br> | ||
| + | |||
|} | |} | ||
<br><br><br> | <br><br><br> | ||
Latest revision as of 03:48, 30 October 2008
|
|---|
|
BioBrick Landing PadsWe will be using landing pads to insert our clock onto the chromosome of E. coli. Noisy behavior has proven detrimental to clock studies throughout the past, and we hope to reduce the noise in our system using landing pads.
BioBrick Landing Pad Polylinker
Landing Pads used for the Sequestilator
|
|---|
 "
"