Team:Rice University/Project
From 2008.igem.org
DavidOuyang (Talk | contribs) (→Recombineering) |
DavidOuyang (Talk | contribs) (→Recombineering) |
||
| Line 20: | Line 20: | ||
=== Roadmap === | === Roadmap === | ||
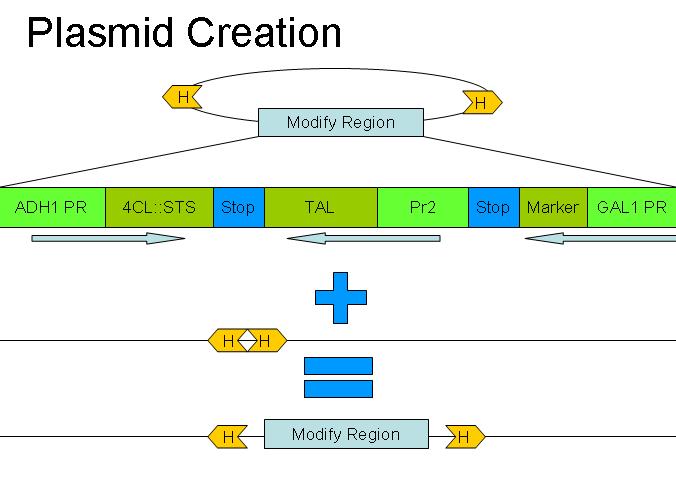
=== Recombineering === | === Recombineering === | ||
| - | [[Image:YeastRecombineeringPlasmid.jpg]] | + | [[Image:YeastRecombineeringPlasmid.jpg|300px|right]] |
=== Experimental Results === | === Experimental Results === | ||
Revision as of 19:46, 24 July 2008
| This year, the Synthetic BiOWLogists are focusing on two new projects. The first project is the creation of BioBeer by incorporation of the resveratrol biosynthetic pathway into the Saccharomyces cerevisiae genome. Resveratrol production during fermentation will be quantified and compared with natural resveratrol sources. The second project is the construction of a novel virus mediated signal amplification system. The initial focus of this project will be to detect and report the presence of an extremely small proportion of a target genotype in a mixed population. |
Contents |
Project 1: BioBeer
Concept
Resveratrol is an unique phytoalexin produced by grapes, peanuts, and knotweed that has known and putative physiological effects. In mice, resveratrol has been linked to anti-inflammatory, anti-cancer, and cardiovascular benefits. Resveratrol has been reported to significantly extend the lifespan of the Saccharomyces cerevisiae. Additionally, resveratrol has been hypothesized to account for the [http://en.wikipedia.org/wiki/French_Paradox French Paradox.]
Using an engineered biosynthetic pathway in Saccharomyces cerevisiae, this project will attempt to create [http://en.wikipedia.org/wiki/marmite marmite] and beer. A proprietary hefeweizen strain was obtained from St. Arnold's [http://www.saintarnold.com/]. This industrial strain is a putative diploid/polyploid strain that is used for fermentation of unfiltered beer.
Pathway Design

Roadmap
Recombineering
Experimental Results
No Taste-testing.
Project 2: Viral Amplification
Background
Cell number and cell type detection is an important area of active interest. Both in cancer detection and monitoring circuit stability, the ability to quantify a small proportion of deviant cells is critical to understand the population mechanics. The ability to distinguish between genotypes will allow researchers to better understand how organisms lose engineered circuits without a selective pressure and allow for earlier treatment of precancerous tumors. Additionally, detection of the presence of unique genotypes in an initially homogeneous population allows for contamination alert.
Fundamental Question: What is the lower limit in cell density and proportion with which we can detect differing cell genotypes and phenotypes?
Foundational Technology:To use an engineered biological system to amplify detection signals.
Concept
By engineering and repackaging temperature-sensitive phage, we plan to implement a signal amplification design that can be generalized to detect unique genotypes within a population.
Design
STF phage
Amber plasmid - tRNA/synthetase
ARFP gene
Detector Circuit
Molecular Biology
Experimental Results
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"