Team:Paris/August 8
From 2008.igem.org
(→Protocol) |
|||
| (45 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | == Minipreps : Plasmid extraction== | + | == Minipreps: Plasmid extraction== |
Extraction of '''pSB3K3 et E0240 in pSB1A2 plasmid''' from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube. | Extraction of '''pSB3K3 et E0240 in pSB1A2 plasmid''' from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube. | ||
| Line 23: | Line 23: | ||
|} | |} | ||
<br> | <br> | ||
| - | |||
== '''Amplification of Genes of interest (OmpR, EnvZ, FlhDC)'''== | == '''Amplification of Genes of interest (OmpR, EnvZ, FlhDC)'''== | ||
| Line 52: | Line 51: | ||
| style="background: #D4E2EF;" | O127 | | style="background: #D4E2EF;" | O127 | ||
| Gene-EnvZ-R | | Gene-EnvZ-R | ||
| - | | | + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTACCCTTCTTTTGTCGTGCCCTGCGCC |
| - | | | + | | 60 |
| | | | ||
|- style="background: #dddddd;" | |- style="background: #dddddd;" | ||
| style="background: #D4E2EF;" | O131 | | style="background: #D4E2EF;" | O131 | ||
| Gene-FlhC-R | | Gene-FlhC-R | ||
| - | | | + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAAACAGCCTGTACTCTCTGTTCATCC |
| - | | | + | | 60 |
| | | | ||
|- style="background: #dddddd;" | |- style="background: #dddddd;" | ||
| Line 76: | Line 75: | ||
| style="background: #D4E2EF;" | O139 | | style="background: #D4E2EF;" | O139 | ||
| Gene-OmpR-R | | Gene-OmpR-R | ||
| - | | | + | | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAGGCCCTTTTCTTGCGCAGCGCTTCT |
| 59 | | 59 | ||
| | | | ||
| Line 82: | Line 81: | ||
| - | * '''Preparation of the templates''' :<br> | + | * '''Preparation of the templates''' :<br>Resuspension of 1 colony in 100µl of water. |
* '''Preparation of PCR mix''' : | * '''Preparation of PCR mix''' : | ||
| - | ''For each | + | ''For each sample,'' |
1 µl dNTP | 1 µl dNTP | ||
| Line 127: | Line 126: | ||
* Program PCR_Screening : Annealing 55°C - Time élongation 1'30" - Number cycle : 29 | * Program PCR_Screening : Annealing 55°C - Time élongation 1'30" - Number cycle : 29 | ||
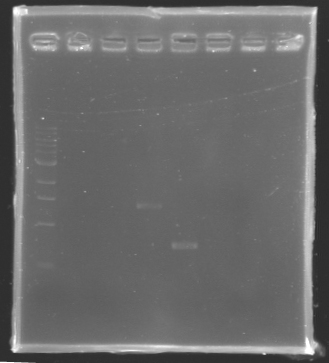
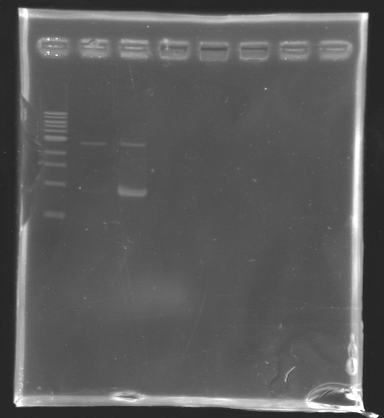
| - | + | ==== '''PCR verification/Analysis''' ==== | |
| - | ==== '''PCR verification/ | + | |
[[Image:KR000129.jpg|thumb|Analysis of PCR product]] | [[Image:KR000129.jpg|thumb|Analysis of PCR product]] | ||
''After the PCR :'' | ''After the PCR :'' | ||
| Line 135: | Line 133: | ||
| - | '''Electrophoresis''' | + | * '''Electrophoresis''' |
| - | + | ladder : 10µl ladder 1 kb | |
| - | + | <br> samples : 3µl of PCR products + 2µl of Loading Dye | |
| - | + | <br> migration 30min at 100V, on a '''1%''' agarose gel | |
| - | * Results : | + | * '''Results :''' |
{| border="1" | {| border="1" | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| Line 178: | Line 176: | ||
| - | '''Quantification of the PCR products purified''' | + | * '''Quantification of the PCR products purified''' |
| - | + | Blank : 2µl of buffer EB + 98µl of water | |
| - | + | <br>Samples : 2µl of PCR purified + 98µl of water. | |
| - | * Results : | + | * '''Results :''' |
{| border="1" | {| border="1" | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| Line 211: | Line 209: | ||
|1.84 | |1.84 | ||
|} | |} | ||
| - | |||
=== Digestion of PCR products === | === Digestion of PCR products === | ||
| Line 268: | Line 265: | ||
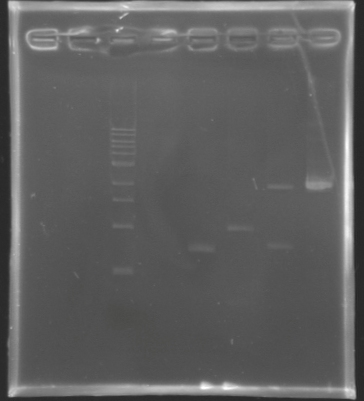
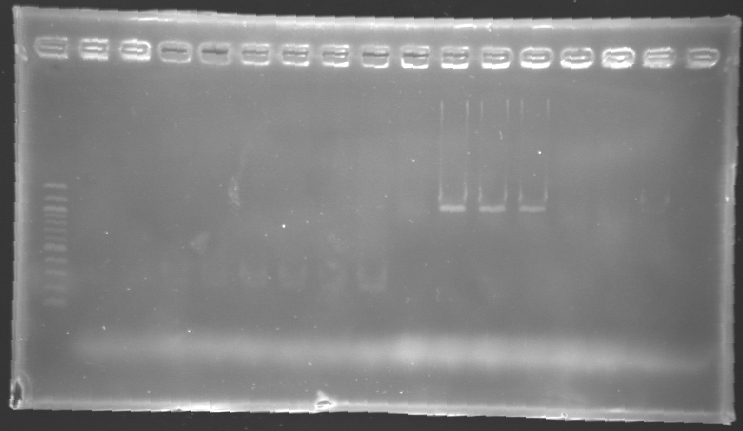
| - | ==== | + | ==== Analysis by electrophoresis ==== |
[[Image:KR000142.jpg|thumb|Analysis of PCR product digestion]] | [[Image:KR000142.jpg|thumb|Analysis of PCR product digestion]] | ||
| - | ''' | + | * '''Conditions''' |
| - | + | ladder : 10µl ladder 1 kb | |
| - | + | <br>samples : 3µl of insert + 2µl of Loading Dye | |
| - | + | <br> migration 30min at 100V, on a '''1%''' agarose gel. | |
| - | * Results : | + | * '''Results :''' |
{| border="1" | {| border="1" | ||
| Line 317: | Line 314: | ||
|} | |} | ||
| - | ==> '''Conclusions : We validate the digestion of the vector and the insert'''. Now we | + | ==> '''Conclusions : We validate the digestion of the vector and the insert'''. Now we are sure, that we don't detect anything for '''FlhDC genes'''. |
| + | == '''The return of the promoters''' == | ||
| + | [[Team:Paris/August_7#Results|Yesterday]], we could not see anything on the electrophoresis after the digestion. Today, we will try the digestion again. | ||
| + | ==='''Protocol=== | ||
| + | {| Border="2" | ||
| + | |align="center"|'''Digestion name''' | ||
| + | |align="center"|'''Template DNA''' | ||
| + | |align="center"|'''Enzymes''' | ||
| + | |align="center"|'''Quantity of DNA used''' | ||
| + | |- | ||
| + | |align="center"|D132 | ||
| + | |align="center"|PCR 124 - pflgA | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|10 µL | ||
| + | |- | ||
| + | |align="center"|D133 | ||
| + | |align="center"|PCR 125 - pflgB | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|10 µL | ||
| + | |- | ||
| + | |align="center"|D134 | ||
| + | |align="center"|PCR 16 - pflhB | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|10 µL | ||
| + | |} | ||
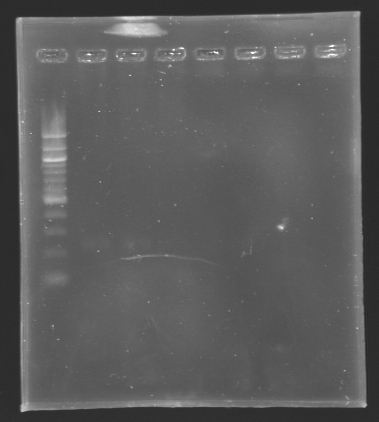
| + | ==='''Results'''=== | ||
| + | [[Image:KR000136.jpg|thumb|Results of the digestion of the promoters]] | ||
| + | To analyze our digestions, we made an electrophoresis. | ||
| + | *Gel : 1.5% Agar | ||
| + | *Ladder : 100 bp | ||
| + | *4µL DNA + 2µL Loading Dye | ||
| + | |||
| + | {| border="1" | ||
| + | |- style="text-align: center;" | ||
| + | |'''Name''' | ||
| + | |'''Promotor''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- style="text-align: center;" | ||
| + | |D132 | ||
| + | |pflgA | ||
| + | |2 | ||
| + | |250 bp | ||
| + | |style="background: #EEAD0E"|<center> 250 bp</center> | ||
| + | |- style="text-align: center;" | ||
| + | |D133 | ||
| + | |pflgB | ||
| + | |3 | ||
| + | |250 bp | ||
| + | |style="background: #EEAD0E"|<center> 250 bp</center> | ||
| + | |- style="text-align: center;" | ||
| + | |D133 | ||
| + | |pflhB | ||
| + | |4 | ||
| + | |249 bp | ||
| + | |style="background: #EEAD0E"|<center> 250 bp</center> | ||
| + | |} | ||
| + | Conclusion | ||
| + | *The bands are not visible on this picture, but with a long exposition time, we managed | ||
| + | to see the bands at their right place. | ||
| + | *The concentration of DNA is low! | ||
| + | We will do the ligation [[Team:Paris/August_9|tomorrow]]. | ||
== '''Transformation with promotors of interest'''== | == '''Transformation with promotors of interest'''== | ||
| Line 428: | Line 487: | ||
|- | |- | ||
|align="center"|'''fluorescence''' | |align="center"|'''fluorescence''' | ||
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
| Line 436: | Line 495: | ||
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
| - | | | + | |style="background: #ff6d73"|red |
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
| Line 452: | Line 511: | ||
|align="center"|no | |align="center"|no | ||
|align="center"|no | |align="center"|no | ||
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
| - | | | + | |style="background: #ff6d73"|red |
|align="center"|no | |align="center"|no | ||
|} | |} | ||
| Line 466: | Line 525: | ||
* '''Mix''' | * '''Mix''' | ||
| - | + | ||
| - | + | 25µl of Quick Load 2X | |
| - | + | <br>1µl of forward primer 10µM | |
| - | + | <br>1µl of reverse primer 10µM | |
| - | + | <br>23µl of water | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
*'''primers used''' | *'''primers used''' | ||
| Line 518: | Line 561: | ||
* Program : Annealing 55°C - Time élongation 1'30" - Number cycle : 29 | * Program : Annealing 55°C - Time élongation 1'30" - Number cycle : 29 | ||
| + | * '''Conditions of electrophoresis ''' | ||
| + | 10µl of ladder 100 pb | ||
| + | <br>10µl of screening PCR | ||
| + | <br>migration ~30min at 100V on '''1,5%''' gel | ||
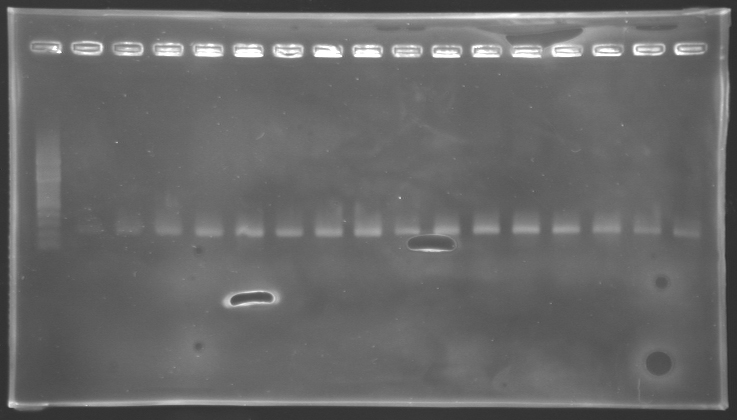
| - | ==== | + | ===='''Results of electrophoresis'''==== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | '''Results of electrophoresis''' | + | |
<br>gel 1 [[Image:KR000143.jpg | gel 1|200px]] | <br>gel 1 [[Image:KR000143.jpg | gel 1|200px]] | ||
| Line 571: | Line 611: | ||
==> '''Conclusion:''' | ==> '''Conclusion:''' | ||
*PCR of pFlgA, pFlgB and pFlhB have succeed, but we always have a problem with pFlhDC probably because of the primers which are not specific. | *PCR of pFlgA, pFlgB and pFlhB have succeed, but we always have a problem with pFlhDC probably because of the primers which are not specific. | ||
| + | |||
| + | |||
| + | =='''Building of the standard measurement plasmid'''== | ||
| + | This morning we MiniPreped E0240 and pSB3K3 which are the two important parts of the standard measurement plasmid. | ||
| + | |||
| + | Before the digestion, we have to determine the DNA concentration of the MiniPreps | ||
| + | |||
| + | ==='''Measurement of DNA concentration'''=== | ||
| + | {| Border="2" | ||
| + | |align="center"|'''Template''' | ||
| + | |align="center"|'''Concentration '''<br>(µg/µL) | ||
| + | |align="center"|'''Ratio DO260/DO280''' | ||
| + | |- | ||
| + | |align="center"|MP 142 <br> pSB3K3 | ||
| + | |align="center"|0.04 | ||
| + | |align="center"|1.76 | ||
| + | |- | ||
| + | |align="center"|MP 143 <br> E0240 | ||
| + | |align="center"|0.16 | ||
| + | |align="center"|1.66 | ||
| + | |} | ||
| + | ==='''Digestion'''=== | ||
| + | ====Protocol==== | ||
| + | {| Border="2" | ||
| + | |align="center"|'''Digestion name''' | ||
| + | |align="center"|'''Template DNA''' | ||
| + | |align="center"|''' Enzymes ''' | ||
| + | |align="center"|'''Volume of DNA''' | ||
| + | |- | ||
| + | |align="center"|D 137 | ||
| + | |align="center"|MP 142 - pSB3K3 | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|25 µL | ||
| + | |- | ||
| + | |align="center"|D 138 | ||
| + | |align="center"|MP 143 - E0240 | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|6.25 µL | ||
| + | |} | ||
| + | |||
| + | * X µL of Template DNA | ||
| + | * Buffer (n°2) 10X : 3µL | ||
| + | * BSA 100X : 0.3µL | ||
| + | * Pure water qsp 30 µL | ||
| + | * 1 µL of each enzyme | ||
| + | |||
| + | * Incubate during about 3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). | ||
| + | |||
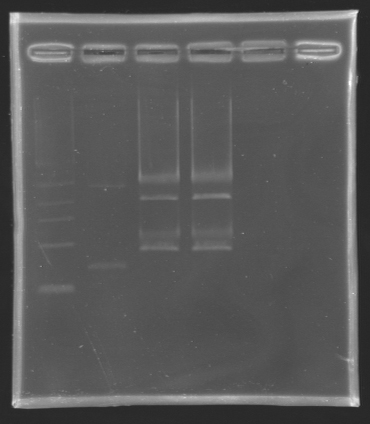
| + | ====Results of the digestion==== | ||
| + | |||
| + | [[Image:KR000131.jpg|thumb|Result of the digestion to build the measurement plasmid]] | ||
| + | |||
| + | '''Electrophoresis settings''' | ||
| + | * Gel : 1 % agar | ||
| + | * 4µL template DNA for D137 | ||
| + | * All the digestion product for D138 because we have to separate two products, the backbone measures 2056 bp and the insert we want to extract is 899 bp long. | ||
| + | * 10µL QuickLoad DNA ladder 1 kb | ||
| + | |||
| + | |||
| + | {| border="1" | ||
| + | |align="center"|'''Name''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- | ||
| + | |align="center"|D 137 | ||
| + | |align="center"|2 | ||
| + | |style="background: #cbff7B"|<center>2727 bp</center> | ||
| + | |align="center"|3000 bp | ||
| + | |- | ||
| + | |align="center"|D 138 | ||
| + | |align="center"|3&4 | ||
| + | |style="background: #cbff7B"|<center>899 bp</center> | ||
| + | |align="center"|~1000 bp | ||
| + | |} | ||
| + | |||
| + | ==='''Gel extraction and DNA purification'''=== | ||
| + | To extract the biobrick E0240 from the gel, we used the standard protocol [[Team:Paris/Notebook/Protocols#Extraction|number 8]].<br> | ||
| + | To purify the DNA we used the standard protocol [[Team:Paris/Notebook/Protocols#Purification_.28Kit_Promega.29|number 10]] | ||
| + | |||
| + | The ligation will be done [[Team:Paris/August_9|tomorrow]]. | ||
| + | |||
| + | =='''Building an other [[Team:Paris/Parts/Plastest|measurement]] plasmid'''== | ||
| + | ==='''PCR to create the special E0240'''=== | ||
| + | ====Protocol==== | ||
| + | PCR Mix for cloning with Taq polymerase | ||
| + | * 25µL Quick-load Mix | ||
| + | * 1µL Oligo F (10µM) | ||
| + | * 1µL Oligo R (10µM) | ||
| + | * 1µL Template DNA ([[Team:Paris/Notebook/Freezer#Plasmids:_MiniPrep_Products|MP 143]]) | ||
| + | * 23µL pure water | ||
| + | |||
| + | We try to build a measurement tool with and without included RBS. | ||
| + | {| Border="2" | ||
| + | |align="center"|'''PCR name''' | ||
| + | |align="center"|'''Template DNA''' | ||
| + | |align="center"|'''Oligo F''' | ||
| + | |align="center"|'''Oligo R''' | ||
| + | |- | ||
| + | |align="center"|RBS + | ||
| + | |align="center"|[[Team:Paris/Notebook/Freezer#Plasmids:_MiniPrep_Products|MP 143]] | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 141]] | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 140]] | ||
| + | |- | ||
| + | |align="center"|RBS - | ||
| + | |align="center"|[[Team:Paris/Notebook/Freezer#Plasmids:_MiniPrep_Products|MP 143]] | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 142]] | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 140]] | ||
| + | |- | ||
| + | |align="center"|T | ||
| + | |align="center"|empty | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 142]] | ||
| + | |align="center"|[[Team:Paris/Notebook/Oligo|O 140]] | ||
| + | |} | ||
| + | |||
| + | '''PCR Program''' | ||
| + | LID 105°C<br> | ||
| + | 1. 95°C 5 min | ||
| + | 2. 95°C 1 min | ||
| + | 3. 60°C 30 sec | ||
| + | 4. 72°C 1 min 30 | ||
| + | 5. go to : 2 rep : 29 | ||
| + | 6. sound : 1 | ||
| + | 7. hold : 10°C | ||
| + | |||
| + | ====Results==== | ||
| + | [[Image:KR000135.jpg|thumb|Results of the PCR]] | ||
| + | '''Electrophoresis settings''' | ||
| + | * Gel : 1 % agar | ||
| + | * 5µL PCR products | ||
| + | * 10µL QuickLoad DNA ladder 1 kb | ||
| + | |||
| + | {| border="1" | ||
| + | |align="center"|'''Name''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- | ||
| + | |align="center"|RBS + | ||
| + | |align="center"|2 | ||
| + | |style="background: #cbff7B"|<center>900 bp</center> | ||
| + | |align="center"|~ 3000 bp <br> ~ 1000 bp (low fluorescence) | ||
| + | |- | ||
| + | |align="center"|RBS - | ||
| + | |align="center"|3 | ||
| + | |style="background: #cbff7B"|<center>881 bp</center> | ||
| + | |align="center"|~ 3000 bp <br> ~ 1000 bp (strong fluo) | ||
| + | |- | ||
| + | |align="center"|T | ||
| + | |align="center"|4 | ||
| + | |style="background: #cbff7B"|<center>nothing</center> | ||
| + | |align="center"|nothing | ||
| + | |} | ||
| + | |||
| + | Conclusion: | ||
| + | - There is DNA that is around 3,000 bp. Actually, it is the template DNA (MP 143), there were too much DNA. | ||
| + | We will have to extract on gel the right piece of DNA. | ||
| + | - RBS - worked better than RBS +. | ||
| + | |||
| + | ====Gel Extraction and DNA purification==== | ||
| + | [[Image:KR000138.jpg|thumb|Results of the PCR]] | ||
| + | '''Electrophoresis settings''' | ||
| + | * Gel : 1 % agar | ||
| + | * All the PCR products (Two bands for each template) | ||
| + | * 10µL QuickLoad DNA ladder 1 kb | ||
| + | |||
| + | Once again we see that the PCR for RBS - worked a lot better than the PCR for RBS +. We will do it again on [[Team:Paris/August_11|monday]]. | ||
| + | |||
| + | To extract RBS - from the gel, we used the standard protocol [[Team:Paris/Notebook/Protocols#Extraction|number 8]].<br> | ||
| + | To purify the DNA we used the standard protocol [[Team:Paris/Notebook/Protocols#Purification_.28Kit_Promega.29|number 10]] | ||
Latest revision as of 13:22, 15 August 2008
|
Minipreps: Plasmid extractionExtraction of pSB3K3 et E0240 in pSB1A2 plasmid from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube.
Amplification of Genes of interest (OmpR, EnvZ, FlhDC)We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR amplificationProtocol
For each sample, 1 µl dNTP
PCR verification/AnalysisAfter the PCR :
ladder : 10µl ladder 1 kb
==> Conclusion : we observed the size expected for the PCR products, but not for FlhDC gene.
Blank : 2µl of buffer EB + 98µl of water
Digestion of PCR productsProtocol :
Analysis by electrophoresis
ladder : 10µl ladder 1 kb
==> Conclusions : We validate the digestion of the vector and the insert. Now we are sure, that we don't detect anything for FlhDC genes. The return of the promotersYesterday, we could not see anything on the electrophoresis after the digestion. Today, we will try the digestion again. Protocol
ResultsTo analyze our digestions, we made an electrophoresis.
Conclusion
*The bands are not visible on this picture, but with a long exposition time, we managed
to see the bands at their right place.
*The concentration of DNA is low!
We will do the ligation tomorrow. Transformation with promotors of interestResults
PCR Screening of Ligation TransformantsUse of 8 clones of Ligation transformants for PCR screening
Protocol of screening PCR
25µl of Quick Load 2X
10µl of ladder 100 pb
Results of electrophoresis
Building of the standard measurement plasmidThis morning we MiniPreped E0240 and pSB3K3 which are the two important parts of the standard measurement plasmid. Before the digestion, we have to determine the DNA concentration of the MiniPreps Measurement of DNA concentration
DigestionProtocol
Results of the digestionElectrophoresis settings
Gel extraction and DNA purificationTo extract the biobrick E0240 from the gel, we used the standard protocol number 8. The ligation will be done tomorrow. Building an other measurement plasmidPCR to create the special E0240ProtocolPCR Mix for cloning with Taq polymerase
We try to build a measurement tool with and without included RBS.
PCR Program LID 105°C ResultsElectrophoresis settings
Conclusion:
- There is DNA that is around 3,000 bp. Actually, it is the template DNA (MP 143), there were too much DNA.
We will have to extract on gel the right piece of DNA.
- RBS - worked better than RBS +.
Gel Extraction and DNA purificationElectrophoresis settings
Once again we see that the PCR for RBS - worked a lot better than the PCR for RBS +. We will do it again on monday. To extract RBS - from the gel, we used the standard protocol number 8. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"