Team:Paris/Modeling/BOB
From 2008.igem.org
(Difference between revisions)
(→Introduction) |
|||
| Line 6: | Line 6: | ||
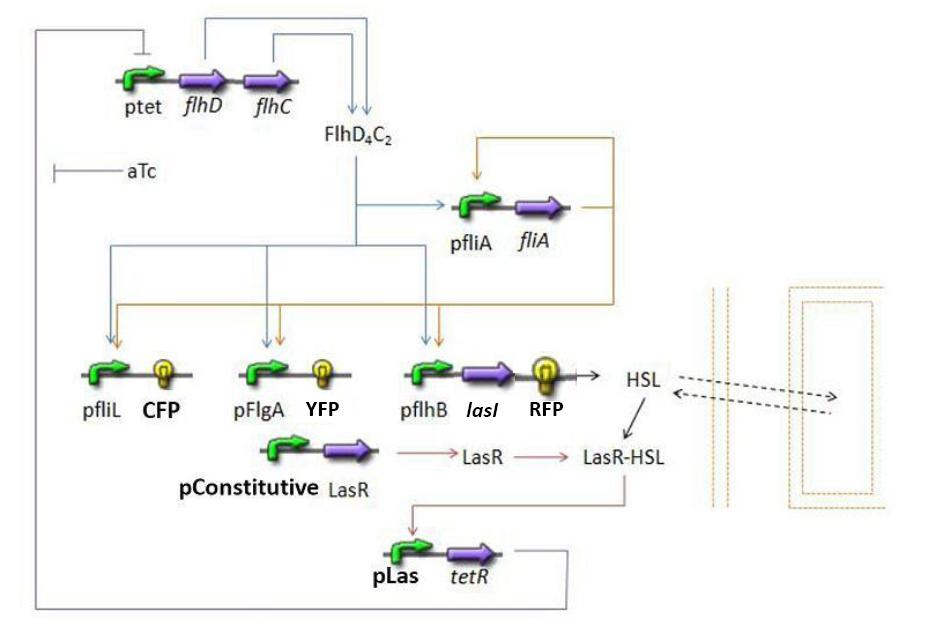
* This preliminary approach is mainly based on a bibliographical work. In fact, it is important to understand that with this approach, we did not intend to design the most accurate model possible. We found essential to choose only parameters that we could determine or control in the wet lab. The main goal was to get quickly a first idea of the way our system could behave. | * This preliminary approach is mainly based on a bibliographical work. In fact, it is important to understand that with this approach, we did not intend to design the most accurate model possible. We found essential to choose only parameters that we could determine or control in the wet lab. The main goal was to get quickly a first idea of the way our system could behave. | ||
* The goal here is to present the differential equations we used in the model. At each step we shall describe why we chose these precise equations, the drawbacks and possible improvements, the parameters involved and eventually a biologically coherent value. | * The goal here is to present the differential equations we used in the model. At each step we shall describe why we chose these precise equations, the drawbacks and possible improvements, the parameters involved and eventually a biologically coherent value. | ||
| + | |||
| + | [[Image:Globalmodel.jpg|600px|center]] | ||
Revision as of 16:01, 27 August 2008
Introduction
|
 "
"