From 2008.igem.org
(Difference between revisions)
|
|
| Line 66: |
Line 66: |
| | <br><br> | | <br><br> |
| | '''Method:''' <br> | | '''Method:''' <br> |
| - | A classical chemostat model using Ordinary Differential Equations was constructed and analyised in terms of sensitivity analysis and simulation of realitstic data. | + | A classical chemostat model using Ordinary Differential Equations was constructed and analyised in terms of sensitivity analysis and simulation of realitstic data.<br><br> |
| - | <br><br> | + | |
| | '''Results:''' <br> | | '''Results:''' <br> |
| - |
| |
| | <br><br> | | <br><br> |
| | </div> | | </div> |
Revision as of 16:32, 11 October 2008
Overview on the modelling framework
This page is meant to give an introduction to the the overall modelling framework we have constructed in order to asses feasibility analysis, temporal scale details and other parameter estimations that regard our project setup. As introduced in the project overview section, four main components can be identified in the deviced mechanism. Accordingly, we divided the modelling framework in four modules that tackles the relative problematics.
The first module is concerned with the analysis of restriction enzymes and their cutting pattern on E.Coli genome, the second module predict the cell's response to the selection pressure and the forced genome reduction from a system point of view (that is, using a genome scale model), the third module addresses issues related to the sensitivity and setting of the chemostat mechanism, the fourth and final module presents the mathematic model of the genetic switch circuit used to control the restriction enzymes expression.
In the table below, you can find a bird-eye view on the four modules, with the most important aspects highlighted. Since we believe that a model is useful only when it answers specific and well-posed questions, this is the first aspect we report in the summary view. Second we briefly report about the modelling method applied. As last, we summarize the results we obtained.
By clicking on each module's title, you can browse the specific module pages containing all the detailed information, such as plots, modelling assumpations and data sources. It is as well possible to download all the data and code (MATLAB source) that we wrote and used in order to generate the results.
Modelling Framework
|
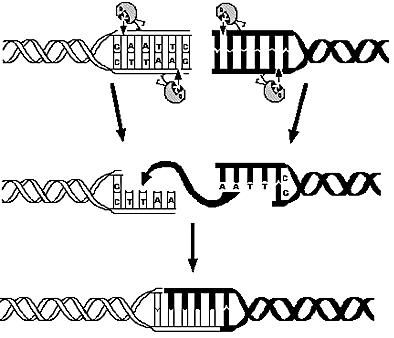
1) Restriction Enzymes Analysis
Questions:
- Which are the available restriction enzymes and cutting patterns?
- How is the distribution of the genes in each fragment related to the frequence of cutting?
- Is it possible to identify a restriction enzyme that optimizes the probability of genome reduced but vital strains?
Method:
E.Coli K12 genome was digested using 713 different restriction enzymes and using annotation information statistical analysis was applied on the fragment patterns.
Results:
The property of a restriction enzymes are all related to its frequency of cutting. The mean number of genes for fragments as well as its variance and the probability of containing essential genes can be derived from that only information.
|
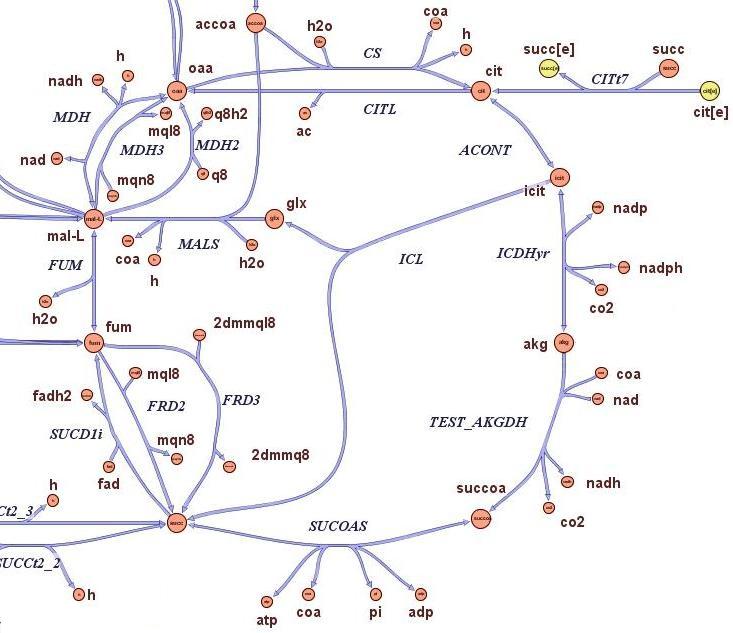
2) Genome Scale Analysis
Questions:
- Is it possible to slow growth of large genome strains by using a thymidine auxotrophyc strain and limiting thymidine feeding?
- What is the best restriction enzyme to be used in order to maximize genome reduction and at the same time vitality (growth rate) of thymidine auxotrophyc strains?
- What is the predicted genome reduction difference if the medium is minimal or very rich in term of nutrients?
- Which are the predicted quantitative differences in terms of growth rate and genome size of strains on which has been applied the selection procedure?
Method:
The state-of-the-art genome scale model for E.Coli iAF1260 (1260 genes included) was modified in order to account for thymidine auxotrophycity, thymidine uptaking limitation, genome reduction and growth on different medium. Stochastic algorithm and flux balance analysis were applied to predict growth rates.
Results:
It is indeed possible to select reduced genome strains using thymidine limitation. The quantification shows that the method is at the border line with the sensitivity of chemostat machinery setup. Predictions shows the possibility of reducing 4% of genes for a minimal medium growing strains and 8% of genese for rich medium growing strains.
|
|
|
|
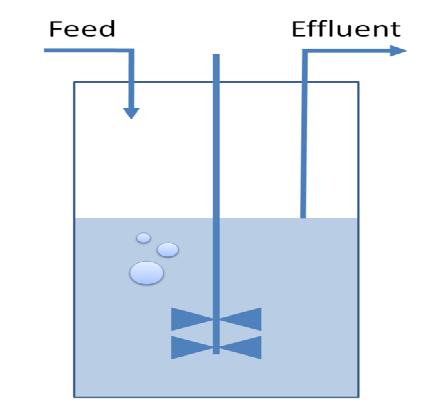
3) Chemostat Selection Method
Questions:
- What are the ideal settings of nutrient concentrations and influx in order to select reduced strains?
- Which is the sensitivity regarding growth rate selection?
- What are the timing parameter in order to induced two subsequent round of restriction enzyme expression?
Method:
A classical chemostat model using Ordinary Differential Equations was constructed and analyised in terms of sensitivity analysis and simulation of realitstic data.
Results:
|
4) Switch Circuit
Questions:
Method:
Results:
|
|
 "
"