|
__september
Sep. 10th 2008
CMV PCR (Sabine)
-PCR with primer for the CMV promotor; as DNA template the CMV+RLuc construct from the Ljubljana group (isolated from the parts collection 2007) was used.
For a 50 µl reaction:
40,4 µl H2O
5 µl buffer (10x)
1,5 µl FWD Primer (15pmol)(
1,5 µl REV Primer (15pmol)
1 µl dNTP (10mM)
0,1 µl DNA template
0,5 µl Pfu Polymerase
The settings for the PCR machine are the following:
1. T=94°C 00:02:00
2. T=94°C 00:00:30
3. T=62°C 00:00:30
4. T=72°C 00:01:00
5. GOTO 2 REP 29
6. T=72°C 00:10:00
7. HOLD 6°C
No product was received.
Sep. 11th 2008
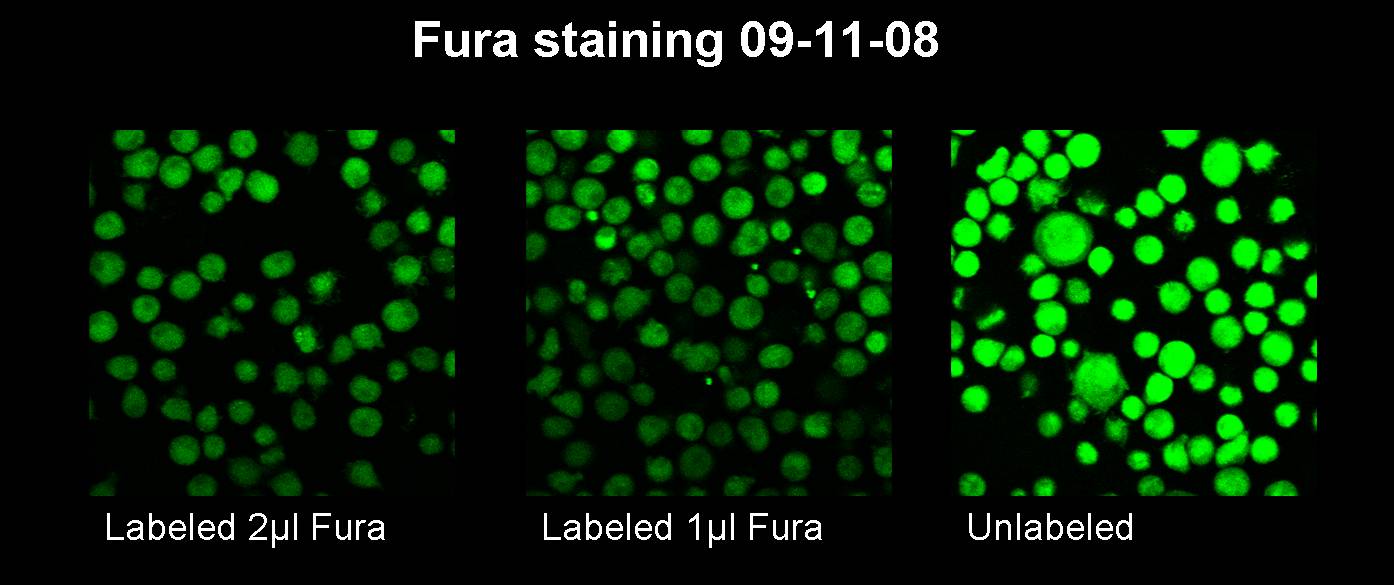
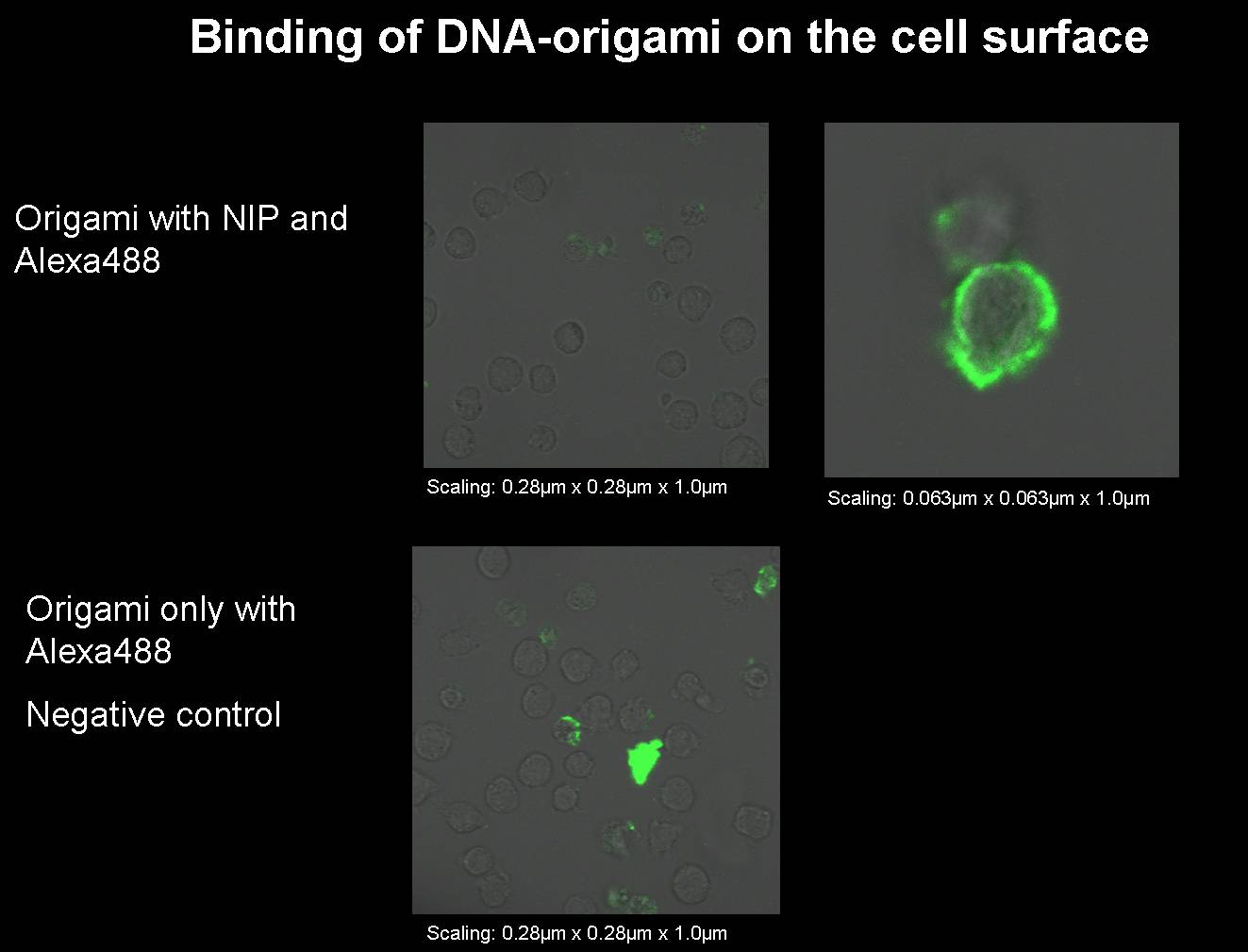
Sep. 12th 2008
1) Origami with NIP and fluorophor for the binding measurement
We had to produce some new origami for our next binding measurements.
- Origami with NIP and fluorophor
- Origami only with fluorophor (without NIP); negative control
see at the protocol from 07-24-2008
2) Origami for the Calciummeasurement
- Origami without NIP (negative control)
see at the protocol from 07-24-2008
To increase the concentration of origami we also made to probes with the double amount
ingredients of the protocol from 07.24.2008
|
Origami with NIP (6x (1:5)) [µl] |
Origami without NIP (6x (1:5)) [µl] |
| Oligos-Pool |
43,68 |
43,68 |
| remainders |
2,4 |
2,4 |
| MgAc |
1 |
1 |
| Phage DNA (448,4 mg/µl) |
33,6 |
33,6 |
| NIP-Oligo |
1,68 |
---- |
| Pool oligo without fluorophor |
0,72 |
0,72 |
| Oligo without NIP |
---- |
1,68 |
3) Master cycler
The origamis were produced in the mastercycler as explained before.
4) Purification of the DNA Origami
Was done as before
5) Digestion of CMV+Rluc (Sabine)
Digestion with EcoRV und FspI (3h at 37°C)
- 5µl plasmid
- 10µl H2O
- 0,5µl enzyme
- 2,5µl buffer (2)
- 0,5µl BSA
File:Verdau CMVRluc EcoRV FspI klein.jpg
6) CMV PCR (Sabine)
-the same PCR with different annealing temperatures using a gradient between 58°C and 62°C (optimal annealing temperature: 62°C)
-again, no products were gained
Sep. 15th 2008
CMV-PCR (Sabine)
-another effort to gain the CMV-Promotor via PCR, this time with different polymerases: Taq Polymerase and a Mix.
-one approach with the complete plasmid and one with the digested plasmid (see digestion of CMV+Rluc)
-products in the approach with taq polymerase as well as in the approach with the Mix. 3 bands from each approach were cut out
Sep. 16th 2008
Gel purification (Sabine)
-gel purification of the PCR products
Digestion of the PCR products and the transfectionvector (Sabine, Kathrin)
-digestion with EcoRI and PstI
-ligation
Sep. 18th 2008
Transformation (Sabine)
-transformation with the ligation product.
Sep. 19th 2008
Transformation (Sabine)
-no colonies on the plates.
Sep. 20th 2008
Digestion of the PCR products and the transfection-vector (Sabine)
-the restriction-enzymes XbaI and SpeI were used
Gel purification and ligation (Sabine)
-digestion of PCR products and vector
Sep. 21st 2008
Transformation of the ligation (Sabine)
-RV 308 cells were transformed with 10 µl of the ligation
Sep. 23rd 2008
1. Transformation of the 4 plasmids of ATG (michael)
-> Signalpeptide
-> Transmembrane domain
-> His-tag
-> Strep-tag
2. Digestion...
Sep. 24th 2008
1. Transformation (michael)
of the Ligations: CMV-Promotor + Vector
proportion: PCR-product (CMV promoter) / vector
-> 6/2
-> 4/4
2. Picking clons of the '4 ATG trafos' (michael)
-> Signalpeptide
-> Transmembrane domain
-> His-tag
-> Strep-tag
after growing glycerinstocks and Miniprep of all 4
3. Picking clons from stock (michael)
-> lipocalin
-> C-GFP and N-GFP
-> C-CFP and N-CFP
-> scFv anti NIP
4. Digestion of the CMV-promoter
Approach Mix: 3/4.1/4.2/4.3/4.4
...
Sep. 25th 2008
Miniprep (normann)
of C-CFP and scFv-anti-NIP
Picking from stocks (normann):
N-CFP, N-GFP, C-CFP, C-GFP
Defrost B12-cells (michael)
and put in new RPMI:
->500ml RPMI
+50ml FCS(10%)
+5ml Hepes(1M)
+5ml L-glutamine(200mM)
+5ml Pen-Strep
+1,75µl ß-Mercaptoethanol(14.3M)
Ligation 2nd try (sabine)
with T4-ligase:
Insert: CMV-Promotor, old and new PCR-Product
Vector: Transfection-Vector fermented with EcoRI/SpeI
-Approaches (for each, old and new PCR-Product): Insert/Vector -> a) 6/2, b) 4/4
Sep. 26th 2008
Transformation of the 'ligation 2nd try' (michael)
Picking clones
4x Signalpeptide
2x Transmembraneregion
Miniprep of: (norman)
-Lipocalin
-Venus split N-GFP
-Venus split C-GFP
-Cerulan split N-CFP
|  "
"


