Team:Paris/Notebook/Protocols
From 2008.igem.org
(→Transformation) |
(→PCR Screening) |
||
| (44 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
* Streaks on plates with LB and the adapted antibiotics to isolate colonies | * Streaks on plates with LB and the adapted antibiotics to isolate colonies | ||
* Incubate O/N at 37°C | * Incubate O/N at 37°C | ||
| - | * Take clone with a | + | * Take clone with a sterile tip and put in 7.5ml LB with adaptated antibiotics (LB have to be prepared before !!!) |
* Will be use for Miniprep and Stock in glycerol | * Will be use for Miniprep and Stock in glycerol | ||
* 2-3 clones isolated by Biobricks | * 2-3 clones isolated by Biobricks | ||
| Line 28: | Line 28: | ||
* Wash QIAprep spin column by adding 0.75 ml Buffer PE and centrifuging for 30–60 s. | * Wash QIAprep spin column by adding 0.75 ml Buffer PE and centrifuging for 30–60 s. | ||
* Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer. | * Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer. | ||
| - | * To elute DNA, place the QIAprep column in a clean 1.5 ml microcentrifuge tube. Add 30 µl Buffer EB or water to the center of each QIAprep spin column, let stand for 1 min, and centrifuge for 1 min. | + | * To elute DNA, place the QIAprep column in a clean 1.5 ml microcentrifuge tube. Add 30 µl Buffer EB or pure water to the center of each QIAprep spin column, let stand for 1 min, and centrifuge for 1 min. |
= Electrophoresis = | = Electrophoresis = | ||
| Line 34: | Line 34: | ||
* Gel : 1% agarose with BET added (5 µL BET for 100 mL TBE) | * Gel : 1% agarose with BET added (5 µL BET for 100 mL TBE) | ||
* 10 µL Quick-Load 1 kb DNA Ladder | * 10 µL Quick-Load 1 kb DNA Ladder | ||
| - | * 2 µL | + | * 2 µL Loading Dye + 3 µL DNA |
| - | + | ||
| - | = Concentration of the Miniprep = | + | = Concentration of the Miniprep or the Midiprep = |
By biophotometry | By biophotometry | ||
| - | * Blank : 55 µL of pure water + 5 µL EB | + | * Blank : 60 µL of pure water or 55 µL of pure water + 5 µL EB |
| - | * Sample : 55 µL of pure water + 5 µL of | + | * Sample : 55 µL of pure water + 5 µL of sample |
Check if the ratio 260/280 is over 1,6 | Check if the ratio 260/280 is over 1,6 | ||
'''Think about the dilution !''' | '''Think about the dilution !''' | ||
| + | |||
| + | If the concentration is to low, concentrate the sample in a vacuum centrifuge. Don't forget to open the tubes! | ||
= Digestion = | = Digestion = | ||
| - | * 1 µg of plasmid / 250 ng of | + | * 1 µg of plasmid / 250 ng of PCR product |
* Buffer (n°2) 10X : 3µL | * Buffer (n°2) 10X : 3µL | ||
* BSA 100X : 0.3µL | * BSA 100X : 0.3µL | ||
| - | |||
* 1 µL of each enzyme | * 1 µL of each enzyme | ||
| + | * Pure water qsp 30 µL | ||
* Incubate during about 2h30-3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). | * Incubate during about 2h30-3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). | ||
= Migration after digestion = | = Migration after digestion = | ||
| - | * Run the whole samples (30 µL) in a ''' | + | * Run the whole samples (30 µL) in a '''1-2% agarose gel with TAE buffer''' (a new one) |
* Run at 50 V until halfway | * Run at 50 V until halfway | ||
* 10 µL of ladder 1kb and 100 pb on every side | * 10 µL of ladder 1kb and 100 pb on every side | ||
| - | * 30 µL of DNA + 6 µL of | + | * 30 µL of DNA + 6 µL of Loading Dye |
| - | '''Separate each | + | '''Separate each sample by an empty well !''' |
= Extraction = | = Extraction = | ||
| - | * For each new extraction it's important to have a new bath of ETB | + | * For each new extraction it's important to have a new bath of TAE Buffer for the run and of ETB for the revelation |
| + | * Use UV at lower intensity and for shorter time as possible | ||
* Use a new blade for each extraction | * Use a new blade for each extraction | ||
| - | * The band weight must be less than | + | * The band weight must be less than 400 mg |
| + | |||
| + | |||
| + | = Antarctic phosphatase = | ||
| + | To avoid autoligation, the vector in a construction has to be treated with phosphatase to dephosphorylate the cut end of the plasmid. The Antarctic phosphatase has the advantage to be easily and totally inactivated by heating. | ||
| + | * After the digestion reaction add Antarctic phosphatase buffer to a final concentration of 1x | ||
| + | * Add 1µL of Antarctic phosphatase for 1 to 5 µg of DNA | ||
| + | * Incubate at 37°C for 15 minutes and then inactivate the phosphatase at 65°C for 5 min | ||
| + | |||
| + | |||
| + | '''Do not use this reaction for the insert or the ligation will not work''' | ||
= Amplification of promoters = | = Amplification of promoters = | ||
| Line 94: | Line 106: | ||
8. hold : 10°C | 8. hold : 10°C | ||
| - | = Purification (PROMEGA kit) = | + | = Purification (PROMEGA or QIAcube or QIAquick kit) = |
== Gel Slice and PCR Product Preparation == | == Gel Slice and PCR Product Preparation == | ||
=== Dissolving the Gel Slice === | === Dissolving the Gel Slice === | ||
| - | * Following electrophoresis, excise DNA band from gel slice in a pre-weighed | + | * Following electrophoresis, excise DNA band from gel slice in a pre-weighed 2 mL microcentrifuge tube. |
* Add 10µL membrane Binding Solution per 10 mg of gel slice. We prefer not to vortex and we incubate at 50-65°C until gel slice is completely dissolved (∼10 min). Quick centrifuge. | * Add 10µL membrane Binding Solution per 10 mg of gel slice. We prefer not to vortex and we incubate at 50-65°C until gel slice is completely dissolved (∼10 min). Quick centrifuge. | ||
| Line 117: | Line 129: | ||
== Elution == | == Elution == | ||
* Carefully transfer Minicolumn to a clean 1.5 mL microcentrifuge tube. | * Carefully transfer Minicolumn to a clean 1.5 mL microcentrifuge tube. | ||
| - | * Add 30 µL | + | * Add 30 µL of pure water or buffer EB (Qiagen). Incubate at room temparature for 1 minute. Centifuge at 16,000 x ''g'' for 1 minute. |
* Discard Minicolumn and store at 4 or -20°C. | * Discard Minicolumn and store at 4 or -20°C. | ||
| - | |||
= Quantification by Electrophoresis = | = Quantification by Electrophoresis = | ||
* Gel : 1.5-2% agarose with BET added (5 µL BET for 100 mL TBE) | * Gel : 1.5-2% agarose with BET added (5 µL BET for 100 mL TBE) | ||
| - | * 10 µL Quick-Load 1 kb DNA Ladder | + | * 10 µL Quick-Load 100pb or 1 kb DNA Ladder |
| - | * 2 µL | + | * 2 µL Loadind Dye + 3 µL DNA |
| - | + | ||
= Ligation = | = Ligation = | ||
| Line 153: | Line 163: | ||
* Spread on appropriate plates | * Spread on appropriate plates | ||
* Incubate O/N at 37°C | * Incubate O/N at 37°C | ||
| + | |||
| + | |||
| + | = LB and LBA = | ||
| + | |||
| + | * LB medium: Dissolve 25mg/L of Luria Broth in purified water. Autoclave the solution. Store at room temperature and take care of any contamination | ||
| + | |||
| + | * LBA medium: Dissolve and gently mix with a magnetic barrel 40mg/L of Luria Broth Agar in purified water. Autoclave the solution. Store at room temperature and take care of any contamination. To make some plates heat the solidified medium with microwave. Wait untill you can hold the bottle in your hands and then add the appropriate antibiotic to a final concentration of 1x. Provide approximatively 20mL per plate in a sterile hood. Returned the plates and store them at +4°C | ||
| + | |||
| + | = Minimum Medium = | ||
| + | |||
| + | *Minimum Medium: | ||
| + | - B1 Vitamin 0.1% | ||
| + | <br>- Uracile 0.2% | ||
| + | <br>- MgSO<sub>4</sub> 2 mM | ||
| + | <br>- CaCl<sub>2</sub> 0.1 mM | ||
| + | <br>- Glycerol 0.4% | ||
| + | <br>- Casamino acids 0.1% | ||
| + | <br>- M9 minimal salts medium 1x | ||
| + | <br>- H<sub>2</sub>O QSP 200 mL | ||
| + | |||
| + | Filter the medium with a cell-culture unit of filtration. If needed add the appropriate antibiotic to a final concentration of 1x | ||
| + | |||
| + | = Soft Agar Medium = | ||
| + | |||
| + | * Add 3.5 to 4g of agar per liter of LB or Minimum Medium. Autoclave the solution. Store at room temperature and take care of any contamination. To make some plates heat the solidified medium with microwave. Wait untill you can hold the bottle in your hands and then add the appropriate antibiotic to a final concentration of 1x. Provide approximatively 20mL per plate in a sterile hood. Returned the plates and store them at +4°C | ||
| + | |||
| + | = 1000x stock antibiotic = | ||
| + | |||
| + | * Chloramphenicol: dissolve 30mg/mL of chloramphenicol in ethanol 100%. Store at room temperature | ||
| + | * Tetracyclin: dissolve 12,5mg/mL of tetracyclin in a 50/50 ethanol/water mix. Store at -20°C | ||
| + | * Ampicillin: dissolve 100mg/mL of ampicillin in water. Filter the solution. Store at +4°C | ||
| + | * Kanamycin: dissolve 100mg/mL of kanamycin in water. Filter the solution. Store at room temperature | ||
= PCR Screening = | = PCR Screening = | ||
Use of 8 clones of Ligation transformants for screening PCR | Use of 8 clones of Ligation transformants for screening PCR | ||
| - | * One | + | * One sterile tip of each clone's colony per PCR tube |
| - | * Use | + | * Use sterile tip to start 7.5mL O/N culture |
'''After''', add | '''After''', add | ||
* 12.5 µL Mix | * 12.5 µL Mix | ||
| - | * 0.5 µL Oligo | + | * 0.5 µL Oligo VF (10µM) |
| - | * 0.5 µL Oligo | + | * 0.5 µL Oligo VR (10µM) |
* 11,5 µL pure water | * 11,5 µL pure water | ||
*negative control : without clone's colony | *negative control : without clone's colony | ||
| Line 179: | Line 221: | ||
6. sound : 1<br> | 6. sound : 1<br> | ||
7. hold : 4°C | 7. hold : 4°C | ||
| - | |||
= Electrophoresis Purification of PCR = | = Electrophoresis Purification of PCR = | ||
| Line 232: | Line 273: | ||
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhDC |
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliA |
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliL |
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlgA |
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlgB |
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhB |
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| Line 255: | Line 296: | ||
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
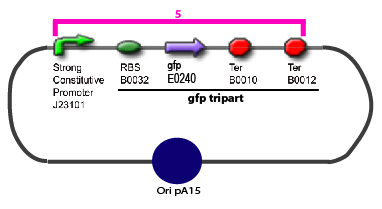
| - | | | + | |GFP E0240 |
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |OmpR* |
| - | | | + | |no |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |EnvZ* |
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |FliA |
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |FlhDC |
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
|colspan="4"|<center>'''Plasmids'''</center> | |colspan="4"|<center>'''Plasmids'''</center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pSB3K3 (ORI p15A) |
| + | |yes | ||
| + | |- style="text-align: center;" | ||
| + | |pSB4T5 (ORI pSC101) | ||
|yes | |yes | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| Line 278: | Line 322: | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
|B0032 | |B0032 | ||
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
|colspan="4"|<center>'''Terminators'''</center> | |colspan="4"|<center>'''Terminators'''</center> | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
|B0010 | |B0010 | ||
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
|B0012 | |B0012 | ||
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
|colspan="4"|<center>'''Bacterial Strains'''</center> | |colspan="4"|<center>'''Bacterial Strains'''</center> | ||
| Line 294: | Line 338: | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
|FliA -/ FlhDC - | |FliA -/ FlhDC - | ||
| - | | | + | |yes |
|- style="text-align: center;" | |- style="text-align: center;" | ||
|FlgM - | |FlgM - | ||
| - | | | + | |yes |
|} | |} | ||
| Line 326: | Line 370: | ||
|yes | |yes | ||
| | | | ||
| - | | | + | | |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| Line 335: | Line 379: | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | | |
| - | | | + | | |
| - | | | + | |GFP E0240 |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhDC |
| - | | | + | |yes |
| - | | | + | |OmpR* |
| - | | | + | |no |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhDC |
| - | | | + | |yes |
| - | | | + | |EnvZ* |
| - | | | + | |yes |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhDC |
| - | | | + | |yes |
| - | | | + | |FliA |
| - | | | + | |yes |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliA |
| - | | | + | |yes |
| - | | | + | |FlhDC |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliA |
| - | | | + | |yes |
| - | | | + | |FliA |
| - | | | + | |yes |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliL |
| - | | | + | |yes |
| - | | | + | |FlhDC |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFliL |
| - | | | + | |yes |
| - | | | + | |FliA |
| - | | | + | |yes |
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlgA |
|yes | |yes | ||
| - | | | + | |FlhDC |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlgA |
| + | |yes | ||
| + | |FliA | ||
|yes | |yes | ||
| - | |||
| - | |||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pflgB |
|yes | |yes | ||
| - | | | + | |FlhDC |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlgB |
| + | |yes | ||
| + | |FliA | ||
|yes | |yes | ||
| - | |||
| - | |||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhB |
|yes | |yes | ||
| - | | | + | |FlhDC |
|yes | |yes | ||
| | | | ||
|- style="text-align: center;" | |- style="text-align: center;" | ||
| - | | | + | |pFlhB |
| + | |yes | ||
| + | |FliA | ||
|yes | |yes | ||
| - | |||
| - | |||
| | | | ||
|} | |} | ||
| Line 422: | Line 466: | ||
= Protocol to make competent bacteria = | = Protocol to make competent bacteria = | ||
| - | 1. Use non competent bacteria (ex: MG1655) stocked in 1.5 mL LB (20% Glycerol): put a | + | 1. Use non competent bacteria (ex: MG1655) stocked in 1.5 mL LB (20% Glycerol): put a sterile tip in the 1.5 mL stock tube and then place it in a 50 mL Falcon with 5 ml LB medium. Over Night culture at 37°C / 300 rpm |
2. 1/100 dilution in LB medium QSP 50 mL in an erlenmeyer of 250 mL | 2. 1/100 dilution in LB medium QSP 50 mL in an erlenmeyer of 250 mL | ||
| Line 449: | Line 493: | ||
9. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl<sub>2</sub> and mix gently the suspension by up and down | 9. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl<sub>2</sub> and mix gently the suspension by up and down | ||
| - | 10. | + | 10. [https://2008.igem.org/Team:Paris/Notebook/Protocols#Transformation Transform] or freeze the competent cells. Freeze the competent cells in 50 µL aliquots in the 0.1M CaCl<sub>2</sub> medium with 15% glycerol. |
| - | 11. | + | 11. After transformation, prepare a [https://2008.igem.org/Team:Paris/Notebook/Protocols#Glycerol_Stocks Glycerol Stock] or/and use the transformed bacteria to study [https://2008.igem.org/Team:Paris/Notebook/Protocols#Study_of_the_doubling_time_of_the_bacteria_population the doubling time of the bacteria population] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
= Study of the doubling time of the bacteria population = | = Study of the doubling time of the bacteria population = | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
* Unfreeze strains from the glycerol stock containing or not the plasmid with a contitutive promoter and a fluorescent reporter gene | * Unfreeze strains from the glycerol stock containing or not the plasmid with a contitutive promoter and a fluorescent reporter gene | ||
| - | * Put a | + | * Put a sterile tip in the 1.5 mL stock tube and then place it in a 50 mL Falcon with 5 ml of [https://2008.igem.org/Team:Paris/Notebook/Protocols#Minimum_Medium minimum medium]. Over Night (16h) culture at 37°C / 300 rpm |
* Measure the OD<sub>600</sub> (Linear Zone Measurement <1.5) | * Measure the OD<sub>600</sub> (Linear Zone Measurement <1.5) | ||
* Dilute the culture medium in the same new medium to obtain an approximative OD<sub>600</sub> of 0.01 (V<sub>final</sub>= 50 mL in a 250 ml erlenmeyer) | * Dilute the culture medium in the same new medium to obtain an approximative OD<sub>600</sub> of 0.01 (V<sub>final</sub>= 50 mL in a 250 ml erlenmeyer) | ||
* Incubate à 37°C / 300 rpm and measure the OD<sub>600</sub> every 20 min | * Incubate à 37°C / 300 rpm and measure the OD<sub>600</sub> every 20 min | ||
* Determine the doubling time population and compare the strains containing or not the plasmid with a contitutive promoter and a fluorescent reporter gene | * Determine the doubling time population and compare the strains containing or not the plasmid with a contitutive promoter and a fluorescent reporter gene | ||
Latest revision as of 12:45, 21 October 2008
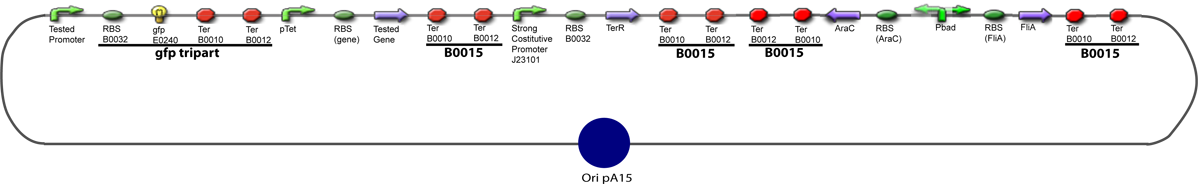
Culture of Stable strain with biobricks 2008
Glycerol Stocks
Minipreps (QIAGEN kit)
ElectrophoresisAn electrophoresis can be done to check if there is Product of Miniprep
Concentration of the Miniprep or the MidiprepBy biophotometry
Check if the ratio 260/280 is over 1,6 Think about the dilution ! If the concentration is to low, concentrate the sample in a vacuum centrifuge. Don't forget to open the tubes! Digestion
Migration after digestion
Separate each sample by an empty well ! Extraction
Antarctic phosphataseTo avoid autoligation, the vector in a construction has to be treated with phosphatase to dephosphorylate the cut end of the plasmid. The Antarctic phosphatase has the advantage to be easily and totally inactivated by heating.
Amplification of promoters(to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments)
For each samples
1 µl dNTP
LID : 105°C Purification (PROMEGA or QIAcube or QIAquick kit)Gel Slice and PCR Product PreparationDissolving the Gel Slice
Processing PCR reactionsFor products above 40 pb
Binding of DNA
Washing
Elution
Quantification by Electrophoresis
Ligation
TransformationUse chemically lab-made competent cells or from a kit (DH5α, TOP10, Mach1)
LB and LBA
Minimum Medium
- B1 Vitamin 0.1%
Filter the medium with a cell-culture unit of filtration. If needed add the appropriate antibiotic to a final concentration of 1x Soft Agar Medium
1000x stock antibiotic
PCR ScreeningUse of 8 clones of Ligation transformants for screening PCR
After, add
Store the tubes on ice waiting for PCR attains 95°C then put the tubes in the machine
LID 105°C Electrophoresis Purification of PCR
Sequencing[http://institut.cochin.inserm.fr/rubric_recherche/Plates-Formes/sequencage_genomique/I18NFolder.2005-02-10.4781618697/page2/fr Sequencing COCHIN]
Promoter Characterization PlanFor theoretical consideration, see estimation of parameters
The same colour coded steps can be perfomed at the same time if elements needed are available The order for treating the colours should of course be:
This table contains the promoters we need to characterize, the transcription factors whose effect on the promoter we want to test, and the plasmid we want to obtain in order to carry out each characterization
Protocol to make competent bacteria1. Use non competent bacteria (ex: MG1655) stocked in 1.5 mL LB (20% Glycerol): put a sterile tip in the 1.5 mL stock tube and then place it in a 50 mL Falcon with 5 ml LB medium. Over Night culture at 37°C / 300 rpm 2. 1/100 dilution in LB medium QSP 50 mL in an erlenmeyer of 250 mL 3. Culture at 37°C / 300 rpm untill OD600 reach 0.6 4. Fast cooling at +4°C by gently shaking the erlen in ice Before: prepare CaCl2 0.1M.
5. Use pre-cooled centrifuge at +4°C. Centrifuge 50 mL of the culture in 50 mL falcon: +4°C / 5 min / 5000 rpm 6. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl2 and mix gently the suspension by up and down 7. Add cold CaCl2 QSP 20 mL and incubate 30 min / +4°C 8. Centrifuge the suspension : +4°C / 5 min / 5000 rpm 9. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl2 and mix gently the suspension by up and down 10. Transform or freeze the competent cells. Freeze the competent cells in 50 µL aliquots in the 0.1M CaCl2 medium with 15% glycerol. 11. After transformation, prepare a Glycerol Stock or/and use the transformed bacteria to study the doubling time of the bacteria population Study of the doubling time of the bacteria population
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"