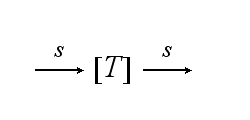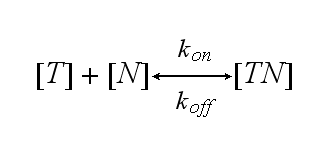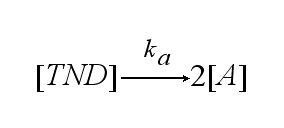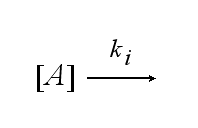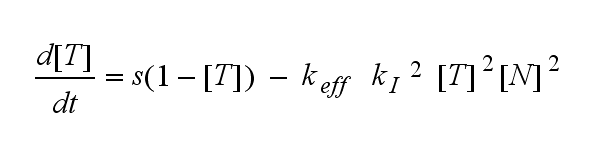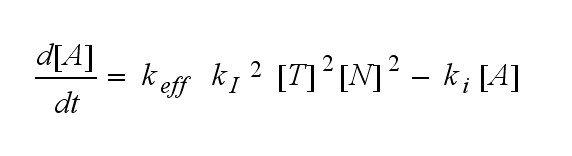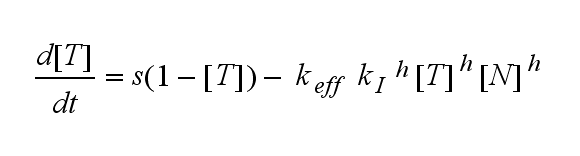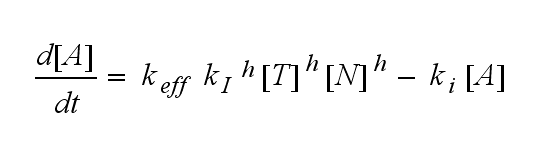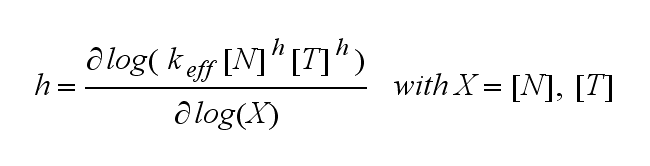Team:Freiburg/Modeling
From 2008.igem.org
| Line 66: | Line 66: | ||
<table> | <table> | ||
<tr> | <tr> | ||
| - | <td>[[image:Freiburg2008_basic1.png|thumb|left| | + | <td>[[image:Freiburg2008_basic1.png|thumb|left|80%]]</td> |
<td>chosen parameters:<br> | <td>chosen parameters:<br> | ||
<font color="#336600"> | <font color="#336600"> | ||
Revision as of 13:19, 27 October 2008
|
Modeling |
_modeling
IntroductionThe dimerization of the extracellular receptor domains is a important necessity for the functionality of our modular receptor system. Presenting the system a stimulus in the form of spatial arranged ligands, the extracellular domains dimerize, thus the corresponding intracellular parts such as the split lactamase halves or split fluorescent proteins complement to measureable output. To analyse the theoretical functionality due to dimerization, first two receptor dimerization models (one T cell receptor model and one general receptor model) are introduced and discussed and then a proper model for the modular receptor system is constructed.
T cell receptor dimerization model I
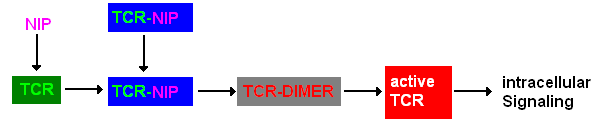
Extracellular signalingA simplified pathway shows the extracellular sequence of TCR activation. After the NIP binding two complexes come together and form a dimer which then leads to activaton of the TCR and further intracellular signaling and T cell activation.
Reaction kinetics
One NIP molecule (N) binds to a TCR (T) with the reaction rate kon or a TCR-NIP complex (TN) dissociates with the reaction rate koff :
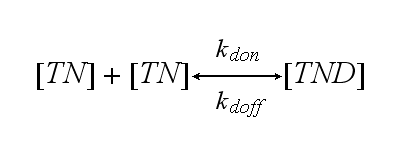
Two TCR-NIP (TN) complexes dimerize to a TCR-NIP dimer (TND) with rate kdon ; the dissociation of a TCR-NIP dimer runs with rate kdoff :
In order to get active TCRs, the TCR-NIP dimer (TND) has to switch into two active TCRs (A) with rate ka :
After activation, the TCR is internalised with rate ki and does not take part anymore in the extracellular signaling :
ODEs derived from the kinetics (Details)In the following equation T represents the free TCR in the T cell membrane where keff is a combination of kdon, kdoff and ka. kI is kon/koff : TCR activity for a set of parametersThe two ODEs above of this first basic model for a set of parameters are solved numerically. They reveal the timecourse of the TCR activity aswell as the one of the unbound TCR.
Extensions: Ultrasensivity and biphasic kineticsUltrasensitivityThe kinetics of the TCR activation can be generalised by substituting the second order kinetic of the ligand N and the receptor T by a parameter h, which then represents the kinetical order of the system. |
 "
"