Team:University of Sheffield /Project
From 2008.igem.org
(→Parts and BioBricks) |
|||
| (114 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | <!-- If you are creating a new page, start cutting here --> | |
| - | + | [[Image:UniShefBanner.jpg|center]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | {| style="color: | + | {| style="color:#888888;background-color:##888888;" cellpadding="5" cellspacing="2" border="2" bordercolor=#888888 width="85%" align="center" |
| - | !align="center"|[[Team:University_of_Sheffield | | + | !align="center"|[[Team:University_of_Sheffield |Introduction]] |
| - | !align="center"|[[Team:University_of_Sheffield / Team | | + | !align="center"|[[Team:University_of_Sheffield /Project|Our project]] |
| - | !align="center"|[[Team:University_of_Sheffield / | + | !align="center"|[[Team:University_of_Sheffield /Modelling|Modelling]] |
| - | !align="center"|[[Team:University_of_Sheffield / | + | !align="center"|[[Team:University_of_Sheffield /Wet Lab|Wet Lab]] |
| - | !align="center"|[[Team:University_of_Sheffield / | + | !align="center"|[[Team:University_of_Sheffield /Lab Books| Our team]] |
| + | !align="center"|[[Team:University_of_Sheffield /Timetable| Timetable]] | ||
| + | !align="center"|[[Team:University_of_Sheffield /Misc| Miscellaneous]] | ||
|} | |} | ||
| - | + | <!-- and finish cutting here! --> | |
| - | + | ||
| - | + | ||
| - | + | =Introduction= | |
| - | + | =Fusion Receptor= | |
| + | ===Overview=== | ||
| + | |||
| + | We hypothesise that expression of receptor protein specific for Vibrio cholerae signalling molecules called autoinducers would confer sensing property on Escherichia coli cell. Because presence of autoinducers in water is associated with cholera pathogens expression of the receptor protein would transform an E.coli into a biosensor. The most straightforward, which does not mean the easiest, way of doing so could be expression of whole signaling pathway in the modified organism. However such an approach leads to a few problems. Firstly, many proteins of downstream signal transduction pathway spanning foreign receptor and DNA have be to introduced into the cell. For example in V.cholera's signalling pathway multiple proteins and small regulatory RNAs are involved. To give response for external stimulus all of them have to be successfully expressed and keep they functionality at satisfactory level. In practice this can be very hard as foreign proteins often tend to form inclusion bodies or their functionality is lowered or lost in a new intracellular conditions they are exposed to. In addition introduction of a whole new pathway can lead to unforeseen consequences for the host cell. | ||
| + | Another method of making bacteria sensitive for external stimulus could be creating fusion receptor protein. In every receptor two elements can be distinguished – receiver and transmitter parts. If two receptors are related closely enough and if they have similar topology fusion of receiver element from one protein with transmitter from the other should give rise to receptor that retains its functionality (Zhu, 2008). Transmitter domain would pass then the signal downstream through a pathway native for modified organism. Such an approach could bring together sensitivity and efficiency of a receptor on one hand and reliability and good level of understanding of signal transduction pathway native to modified organism on the other. | ||
| + | ===Protein analysis=== | ||
| - | + | We want to use this approach in constructing a biosensor monitoring cholera contamination of water. Fusion receptor would consist of two parts, a signal receiver domain from CqsS, protein which is native to V. cholera and binds to CAI-1 molecules (autoinducers synthesised by V.cholerae) and signal conveyer domain of BarA which is a protein present on the surface of E.coli cell and that passes signal downstream to DNA. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Our choice of BarA protein was driven by similarity of those receptors. Both of them are multisubunit molecules of similar topology and use the same method of signal transduction – they belong to Two Component Regulatory System (TCRS). Such a system includes two cooperating proteins a histidine kinase located in the membrane and its partner response regulator. Histidine kinase detects signal from the environment and autophosphorylates itself at the His residue at its cytoplasmic surface. The phosphate group is then passed onto aspartate residue on a response regulator that interacts directly with the DNA or relays signal further on. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Both CqsS and BarA are histidine kinases. Their analysis reveals that they contain sequences that belongs to the same superfamilies - HATPase_c and REC. 3D structures of those proteins haven not been established yet however multisequence alignment and comparison with proteins and protein's domains belonging to the same families revealed conserved structural patterns and probable similar topology. Their function and mechanism of action is most likely conserved. From there we deduced that we should combine N-terminal domain of CqsS protein that is responsible for recognition of autoinducers and autophosphorylation and C-terminal domain of BarA that accepts phosphate and relays it through aspartate downstream. | |
| - | < | + | |
| + | ===Final remarks=== | ||
| + | |||
| + | Big advantage is that we know exactly how the E.coli's native pathway we are employing works. Introduction of reporter system (e.g. GFP) within the site of DNA positively regulated by BarA should show if the stimulus we are sensing for is present in the environment. Shortly if E.coli shines water is contaminated by cholera. | ||
| + | This could be general approach used | ||
| + | |||
| + | Few questions have to be addressed. Firstly, what is the concentration of autoinducers in water that corresponds to amount of pathogens recognised as contamination and if this amount high enough to trigger the response in the receptor. Second question asks how fusion would decrease efficiency of the receptor. Answering these questions is crucial to assess if the machine based on fusion receptor would be of any practical use. | ||
| + | |||
| + | We think that fusing two domains from two different but similar receptors to build a new one could be a general approach to embrace during making biosensors that are based on a foreign receptor. | ||
| + | |||
| + | |||
| + | |||
| + | =The Native E.coli Pathway= | ||
| + | |||
| + | ==Downstream the Fusion Kinase== | ||
| + | |||
| + | The sensing event triggers the phosphorilation of the CqsS-BarA and stimulates the transfer of phosphate group downstream the signaling cascade. Naturally, E.coli passes the phosphate group from BarA to its response regulator UvrY. This particular protein then binds to DNA triggering the transcription of small regulatory RNA called CsrB - Carbon Storage Regulator B. The most complicated part of the pathway is that CsrB inhibits CsrA. CsrA in turn inhibits a whole set of genes depicted in the table on the left ( plus represents activation, minus - inhibition). | ||
| + | |||
| + | One of the genes that CsrA inhibits is responsible for biofilm adhesion protein formation - PGA. This particular operon was our target for reporter molecule insertion. More details about ''pgaABCD'' operon can be found below. | ||
| + | |||
| + | Reference for the images: [[Team:University_of_Sheffield /References#2:_Cerca.2C_N._and_Jefferson.2C_K._K | [2]]] | ||
| + | <div align=center> | ||
| + | [[Image:BarA_Pathway.png|350px|BarA Pathway in E.coli]][[Image:UvrY_Target_Genes.png|300px|UvrY Target Genes]] | ||
<br> | <br> | ||
| - | < | + | </div> |
| - | == | + | =Fluorescent Reporter= |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ===Introduction=== | ||
| + | The initial plan was to introduce a Green Fluorescent reporter molecule under one of the genes controlled by small regulatory RNA called CsrA. So as not to reduce the viability of the cell as well as to avoid any possible “noise” in the engineered system, the gene choice required several attempts. | ||
| + | ===Re-usable testing kit=== | ||
| - | + | The plan to have a micro – chip for sensing biological water contamination ideally would include a possibility of reusing it. Thus, the aim of our project was to use GFP with an LVA tag (AANDENYALAA). This would mean that the GFP would have an amino acid tag targeted by housekeeping proteases and would be degraded within 45 minutes once there is no more stimulation for gene expression. This in turn would mean that the E.coli system sensing V.cholerae presence in water would stop fluorescing once the concentration of CAI-1 molecules decreased up to tolerable level.[[Team:University_of_Sheffield /References#2:_Cerca.2C_N._and_Jefferson.2C_K._K | [2]]] | |
| - | + | The idea to include degradable GFP in our project was taken from the Andersen, J.B et al. (1998) New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria. Applied and Environmental Microbiology. | |
| - | + | ||
| - | + | ||
| + | ===''pgaABCD'' operon control=== | ||
| + | This particular four - gene locus in E.coli is responsible for synthesis of the biofilm adhesion protein called PGA. As this is a virulence factor, i.e. non-essential for growth in supplemented media, a possible insertion of GFP and possible disruption the operon would not cause lethal effects to our cells. | ||
| + | As far as it is known, the ''pgaABCD'' genes are subject to post-translational inhibition by the small regulatory RNA CsrA as well as activation by the cation-responsive regulatory protein NhaR. The first fact is to our advantage. In theory, once the binding of the CAI-1 to fusion kinase occurred, the CsrA inhibition would be releaved and the GFP would be expressed . Also, according to research it has been proven that the ''csrA '' deletion mutant had an impressive 3000 fold increase compared to the wild type. | ||
| + | Possible “noise” in the system | ||
| - | + | As mentioned before, one of the factors to be taken into account prior choosing the gene within which the GFP would be inserted is the interference or “noise” in the system. This possibility might have caused background fluorescence or GFP expression independent from the sensing event of our interest. | |
| - | + | ||
| - | | | + | The reasons underlying this “noise” are listed below: |
| + | *High salt concentrations (NhaR dependent) | ||
| + | *Alkaline pH (NhaR dependent) | ||
| + | *Ethanol ( NhaR dependent) | ||
| + | *Glucose presence in the media (Independent from both CsrA and NhaR) | ||
| + | |||
| + | The noise in the system appears only if the cells are grown at extreme conditions. However, if the growth media contains only 1% glucose, 1 % ethanol, 1 % NaCl the levels of the pgaABCD transcript should not be elevated by any other factors except CsrA. On the other hand, if additional NaCl or glucose are added to the medium it might cause significant increase in the ''pgaABCD'' mRNA appearance [2] | ||
| + | |||
| + | ===Target of insertion=== | ||
| + | |||
| + | As the ''pgaABCD'' is a four-gene locus another question that arose was the actual place within the operon where the insertion could take place. Another obstacle was that genes ''b, c'' and'' d'' overlap with each other. | ||
| + | |||
| + | One would theoretically suggest insertion under the promoter sequence, before the actual gene'' a'' . However, the very high inhibition with CsrA is actually due to 6 binding sites, one of which overlaps with gene ''a'' formyl-methionine codon. | ||
| + | |||
| + | Thus, the only possible way of inserting GFP, with minimal disruption of the whole operon, is at the end of gene ''a'' sequence just before the termination codon. This would give a high level of control by CsrA, as it is just after the CsrA regulatory binding sites, and would hopefully prevent extensive damage to genes ''b,c'' and ''d'' expression. | ||
| + | |||
| + | =Cholera Autoinducers= | ||
| + | Cholera autoinducer (CAI) is a signaling molecule. It is unusual from most pathogen autoinducers as it actually inhibits virulence factor production at high concentration. This is because of the nature of the disease ''V.Cholerae'' causes. The inhibition of virulence factors allows the bacteria to be flushed from the host body into the environment. | ||
| + | |||
| + | |||
| + | CAI is used naturally only by ''V.Cholerae''. This makes it very useful in our bacterial sensor, because unlike other autoinducers, such as AI-2, which are used by many bacteria, this allows us to know bacteria species detected. | ||
| + | |||
| + | CAI is a short ketone with a hydroxyl group. The image below is the different CAI molecules (3 isomer pairs) | ||
| + | |||
| + | [[Image:CAI-shef08.JPG|300px|center|]] | ||
| + | |||
| + | =The Microchip Theory= | ||
| + | [[image:Microchip_diagram.png|450px|right|Our grand master microchip plan.]] | ||
| + | |||
| + | Our idea originated with the concept of a microchip that would test water for pathogen concentration. Whilst one summer wouldn’t allow us to actually build it, we still had an impression as to what it would look like. | ||
| + | ===How?=== | ||
| + | The target was for a cheap, reusable, quick and non-technical piece of equipment. At first, the microchip really was micro, and would require a microscope to look at, but in areas of natural disaster or extreme poverty this wasn’t viable. So we planned and hand-held size chip, with a number of ‘wells’ or ‘chambers’ built into it. Each chamber would contain a different pathogen-sensing E.coli colony. This presented other problems into how to keep the E.coli alive – could they survive if attached physically to the chamber? Or would they have to be frozen, or suspended in media? Frozen seemed the most likely, but not ideal, but proper design could be implemented later. | ||
| + | Then the idea was to take a sample of water, possibly contaminated, and place it on the chip. The contaminants would interact with the specially designed E.coli, and those activated by contaminants would produce a distinctive glow visible to the naked eye. The chamber that glows corresponds to an unhealthy level of a particular pathogen in the water sample. | ||
| + | |||
| + | The reusable aspect came from the idea that disposable equipment is environmentally unfriendly. However, it could be cheaper than a reusable option, so if out plan were to be properly carried out, both chips should be developed to provide options. To make the chip reusable however, LVA tags are attached to the GFP in the DNA code, which attracts a housekeeping protease to cleave the GFP more rapidly than if left to degrade in the cell alone. A short burst of glow would be visible in cell, which would disappear by the time the chip dries out/the water passes through it. | ||
| + | ===Any takers?=== | ||
| + | This approach requires absolutely no technical knowledge, and could be ideal for its purposes. A future iGEM project anyone? | ||
| + | |||
| + | |||
| + | =Parts and BioBricks= | ||
| + | |||
| + | After various attempts to use the Biobrick booklet for transformation procedures needed for implied project ideas, we came to conclusion that the '''booklet''' shipped was actually '''faulty'''. Tests then proved this to be true. Thus the required BioBrick vector for submission of our designed parts was impossible to use from the booklet we were provided. Unfortunately, we ascertained this too late to receive variance approval. Which means we have BioBricks built but no way to send/enter them. | ||
| + | |||
| + | |||
| + | The BioBricks we made, however, were: | ||
| + | |||
| + | *1.) CsrA, with promoter | ||
| + | *2.) CsrA, without promoter | ||
| + | *3.) CsrB, with promoter | ||
| + | *4.) CsrB, without promoter | ||
| + | *5.) UvrY, with promoter | ||
| + | *6.) UvrY, without promoter | ||
| + | *7.) CsqS | ||
| + | *8.) CqsA | ||
| + | *9.) BarA | ||
| + | *10.) Red recombinase system! | ||
| + | *11.) pKD46 Ori site - temperature sensitive | ||
| + | *12.) FLP recombinase | ||
| + | |||
| + | =Acknowledgements= | ||
| + | |||
| + | We would like to thank to everyone who helped us on our way. That is to''' Prof. Visakan Kadirkamanathan''' whose excitement about our | ||
| + | venture made us believe that it is possible. '''Prof. Philip Wright''' for providing us with a fine lab space in the Sheffield Bioincubator. '''Dr Catherine Biggs''' for supporting us with her knowledge and advice.''' Prof. David Hornby''' and his lab for being constant source of practical help. '''Esther Karunakaran''' was playing role of our advisor. She was a patient tutor that guided us through our project. ''' Prof. Jeff Green''' and''' Prof David Rice''' for an advice regarding theoretical aspects of our project. '''Dr Graham Stafford''', '''Bassler Lab''' and '''Ojar Melfors''' for providing us biological material for our experiments. All members of''' ChELSI group''', especially '''Joy''', for their constant advice on every day basis. '''Dave Wengraf''' who knows all legal aspect of science helped us with paper work and got us VAT exemptions. '''Prof. Robert Pool''' and '''Prof. Dominic Shellard''' for providing generous contribution toward our travel to iGEM Jamboree. '''IDT DNA''' for their sponsorship. '''IChemE''', '''BioFusion''' and University of Sheffield '''Union of Students''' for supporting us financially. Without you this project would be much harder, if not impossible to carry out! | ||
| + | |||
| + | =References= | ||
| + | |||
| + | |||
| + | *1.) Andersen, J.B et al. ('''1998''') New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria. '' Applied and Environmental Microbiology'' | ||
| + | |||
| + | * | ||
| + | |||
| + | *2.) Cerca, N. and Jefferson, K. K. ('''2008''') Effect of Growth Conditions On Poly-N-acetylglucosamine expression and Biofilm Formation in Escherichia coli | ||
| + | |||
| + | *3.) Goller, C. et al. ( '''2006''') The Cation-Responsive Protein NhaR of ''Excherichia coli'' Activates ''pgaABCD'' Transcription, Required for Production of the Biofilm Adhesin Poly-Beta-1, 6 - N - D - Glucosamine. ''Journal of Bacteriology'' | ||
| + | |||
| + | *4.)Datsenko & Wanner, ('''2000'''), One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products | ||
| + | |||
| + | *5.) Higgins, Bassler et al, ('''2007'''), The major ''Vibrio cholerae'' autoinducer and its role in virulence factor production | ||
| + | |||
| + | *6.) Hammer & Bassler, ('''2007'''), Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic ''Vibrio cholerae'' | ||
| + | |||
| + | *7.) Jun Zhu, Melissa B. Miller, et al, ('''2001'''), Quorum-sensing regulators control virulence gene expression in ''Vibrio cholerae | ||
| + | '' | ||
| + | *8.)Tomenius, Pernestig et al, ('''2005'''), Genetic and functional characterization of the E.coli BarA-UvrY Two-componant system | ||
| + | |||
| + | *9.) Suzuki et al, ('''2002'''), Regulatory Circuitry of thr CsrA/CrsB and BarA/UvrY systems of E.coli | ||
| + | |||
| + | *10.) Sahu, Acharya et al, ('''2003'''), The bacterial adaptive response gene, barA, encodes a novel conserved istidine kinase regulatory switch for adaptation and modulation of metabolism in E.coli | ||
| + | |||
| + | *11) Sabnis, A. N. et al., ('''1995'''), Pleiotropic Regulation of Central Carbohydrate Metabolism in Escherichia coli via the Gene ''csrA'' | ||
| + | |||
| + | *12) Ytoh, J. et al., ('''2006'''), Roles of pgaABCD Genes in Synthesis, Modification, and Export of the Escherichia coli Biofilm Adhesin Poly-beta-1,6-N-Acetyl-D-Glucosamine | ||
| + | |||
| + | *13) Yang., H. et al. , ('''1995'''), Coordinate Genetic Regulation of Glycogen Catabolism and Biosynthesis in scherichia coli via the CsrA Gene Product | ||
| + | |||
| + | *14) Romeo, T. ('''1998'''), Global Regulation by the small RNA-binding protein CsrA and non-coding RNA molecule CsrB | ||
| + | |||
| + | *15) Wang, X. et al. ('''2005'''), CsrA post-transcriptionally represses pgaABCD,responsible for synthesis of a iofilm polysaccharide adhesin of ''Escherichia coli'' | ||
| + | |||
| + | *16) Romeo, T. et al. ('''1995'''), Identification and Molecular Characterization of ''csrA'', a Pleiotropic Gene from Escherichia coli That Affects Glycogen Biosynthesis, Gluconeogenesis, Cell Size, and Surface Properties | ||
| + | |||
| + | *17) Pernestig, A. K. et al. ('''2002'''), The Escherichia coli BarA-UvrY Two-Component System Is Needed for Efficient Switching between Glycolytic and Gluconeogenic Carbon Sources | ||
| + | |||
| + | *18) Pernestig, A. K. et al. ('''2000'''), Identification of UvrY as the Cognate Response Regulator for the BarA Sensor Kinase in '' Escherichia coli'' | ||
| + | |||
| + | *19) Mondragon, V. et al. ('''2006'''), pH-Dependent Activation of the BarA-UvrY Two-Component System in ''Escherichia coli'' | ||
| + | |||
| + | *20) Rogov, V. V. et al. ('''2006'''), A New Structural Domain in the ''Escherichia coli'' RcsC Hybrid Sensor Kinase onnects Histidine Kinase and Phosphoreceiver Domains | ||
| + | |||
| + | *21) Tomenius, H. et al. ( '''2006'''), The ''Escherichia coli'' BarA-UvrY two-component system is a virulence determinant in the urinary tract | ||
| + | |||
| + | *22) Zhu, Y. and Inouye, M. ('''2004'''), The HAMP Linker in Histidine Kinase Dimeric Receptors Is Critical for Symmetric Transmembrane Signal Transduction | ||
| + | |||
| + | *23) Zhu, y. and Inouye, M. ('''2008'''), Analysis of the Role of the EnvZ Linker Region in Signal Transduction Using a Chimeric Tar/EnvZ Receptor Protein, Tez1 | ||
| + | |||
| + | *24) Walters, M. and Sperandio, V. ('''2006'''), Quorum sensing in ''Escherichia coli'' and ''Salmonella'' | ||
| + | |||
| + | *25) Gonzales, J. E. and Keshavan, N. D. ('''2006'''), Messing with Bacterial Quorum Sensing | ||
| + | |||
| + | *26) Haseltine, E. L. and Arnold, F. H. ('''2007'''), Implications of Rewiring Bacterial Quorum Sensing | ||
| + | |||
| + | *27) Alur, J. et al. ('''2002'''), Modeling and Analyzing Biomolecular Networks | ||
| + | |||
| + | *28) Dunlap, P. V. ('''1999'''), Quorum Regulation of Luminescence in ''Vibrio fischeri'' | ||
| + | |||
| + | *29) Sveningsen, S. L. et al. ('''2008'''), A negative feedback loop involving small RNAs accelerates ''Vibrio cholerae’s'' transition out of quorum-sensing mode | ||
| + | |||
| + | *30) De Silva, R. S. et al. ('''2007'''), Crystal Structure of the ''Vibrio cholerae'' Quorum-Sensing Regulatory Protein HapR | ||
| + | |||
| + | *31) Lenz, D. H. et al. ('''2005'''), CsrA and three redundant small RNAs regulate quorum sensing in ''Vibrio cholerae'' | ||
| + | |||
| + | *32) Heidelberg, J. F. et al. ('''2000'''), DNA sequence of both chromosomes of the cholera pathogen ''Vibrio cholerae | ||
| + | '' | ||
| + | |||
| + | *33) Neidtich, M. B. et al. ('''2006'''), Ligand-Induced Asymmetry in Histidine Sensor Kinase Complex Regulates Quorum Sensing | ||
| + | |||
| + | *34) Matson, J. S. et al. ('''2007'''), Regulatory Networks Controlling ''Vibrio cholerae'' Virulence Gene Expression | ||
| + | |||
| + | *35) Hammer, B. K. and Bassler, B. L. ('''2003'''), Quorum sensing controls biofilm formation in ''Vibrio cholerae'' | ||
| + | |||
| + | *36) Hammer, B. K. and Bassler, B. L. ('''2007'''), Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic ''Vibrio cholerae'' | ||
| + | |||
| + | *37) Lin, W. et al. ('''2005'''), Requirements for ''Vibrio cholerae'' HapR Binding and Transcriptional Repression at the ''hapR'' Promoter Are Distinct from Those at the aphA Promoter | ||
| + | |||
| + | *38) Lenz, D. H. et al. ('''2004'''), The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in ''Vibrio harveyi'' and ''Vibrio cholerae'' | ||
| + | |||
| + | *40) Hoch, J. A. and Varughese, K. J. ('''2001'''), Keeping Signals Straight in Phosphorelay Signal Transduction | ||
| + | |||
| + | *41) McLean, R. J. C. et al. ('''2004'''), A simple screening protocol for the identification of quorum signal antagonists | ||
| + | |||
| + | *42) McClean, K. H. et al. ('''1997'''), Quorum sensing and ''Chromobacterium violaceum'' : exploitation of violacein | ||
| + | production and inhibition for the detection of N-acylhomoserine lactones | ||
| + | |||
| + | *43) Martinko, J. M., Madigan, M. T. ('''2006''') Brock Microbiology of Microorganisms. Pearson Benjamin Cummings, San Fransisco. | ||
| + | |||
| + | *43) Vendeville, A. et al. ('''2005'''), Making‘SENSE’ Of Metabolism:Autoinducer!2, LuxS and Pathogenic Bacteria | ||
| + | |||
| + | *44) Water, C. M. and Bassler, B. L. ('''2005'''), Quorum Sensing: Cell-to-Cell Communication in Bacteria | ||
Latest revision as of 01:07, 29 October 2008
| Introduction | Our project | Modelling | Wet Lab | Our team | Timetable | Miscellaneous |
|---|
Contents |
Introduction
Fusion Receptor
Overview
We hypothesise that expression of receptor protein specific for Vibrio cholerae signalling molecules called autoinducers would confer sensing property on Escherichia coli cell. Because presence of autoinducers in water is associated with cholera pathogens expression of the receptor protein would transform an E.coli into a biosensor. The most straightforward, which does not mean the easiest, way of doing so could be expression of whole signaling pathway in the modified organism. However such an approach leads to a few problems. Firstly, many proteins of downstream signal transduction pathway spanning foreign receptor and DNA have be to introduced into the cell. For example in V.cholera's signalling pathway multiple proteins and small regulatory RNAs are involved. To give response for external stimulus all of them have to be successfully expressed and keep they functionality at satisfactory level. In practice this can be very hard as foreign proteins often tend to form inclusion bodies or their functionality is lowered or lost in a new intracellular conditions they are exposed to. In addition introduction of a whole new pathway can lead to unforeseen consequences for the host cell.
Another method of making bacteria sensitive for external stimulus could be creating fusion receptor protein. In every receptor two elements can be distinguished – receiver and transmitter parts. If two receptors are related closely enough and if they have similar topology fusion of receiver element from one protein with transmitter from the other should give rise to receptor that retains its functionality (Zhu, 2008). Transmitter domain would pass then the signal downstream through a pathway native for modified organism. Such an approach could bring together sensitivity and efficiency of a receptor on one hand and reliability and good level of understanding of signal transduction pathway native to modified organism on the other.
Protein analysis
We want to use this approach in constructing a biosensor monitoring cholera contamination of water. Fusion receptor would consist of two parts, a signal receiver domain from CqsS, protein which is native to V. cholera and binds to CAI-1 molecules (autoinducers synthesised by V.cholerae) and signal conveyer domain of BarA which is a protein present on the surface of E.coli cell and that passes signal downstream to DNA.
Our choice of BarA protein was driven by similarity of those receptors. Both of them are multisubunit molecules of similar topology and use the same method of signal transduction – they belong to Two Component Regulatory System (TCRS). Such a system includes two cooperating proteins a histidine kinase located in the membrane and its partner response regulator. Histidine kinase detects signal from the environment and autophosphorylates itself at the His residue at its cytoplasmic surface. The phosphate group is then passed onto aspartate residue on a response regulator that interacts directly with the DNA or relays signal further on.
Both CqsS and BarA are histidine kinases. Their analysis reveals that they contain sequences that belongs to the same superfamilies - HATPase_c and REC. 3D structures of those proteins haven not been established yet however multisequence alignment and comparison with proteins and protein's domains belonging to the same families revealed conserved structural patterns and probable similar topology. Their function and mechanism of action is most likely conserved. From there we deduced that we should combine N-terminal domain of CqsS protein that is responsible for recognition of autoinducers and autophosphorylation and C-terminal domain of BarA that accepts phosphate and relays it through aspartate downstream.
Final remarks
Big advantage is that we know exactly how the E.coli's native pathway we are employing works. Introduction of reporter system (e.g. GFP) within the site of DNA positively regulated by BarA should show if the stimulus we are sensing for is present in the environment. Shortly if E.coli shines water is contaminated by cholera. This could be general approach used
Few questions have to be addressed. Firstly, what is the concentration of autoinducers in water that corresponds to amount of pathogens recognised as contamination and if this amount high enough to trigger the response in the receptor. Second question asks how fusion would decrease efficiency of the receptor. Answering these questions is crucial to assess if the machine based on fusion receptor would be of any practical use.
We think that fusing two domains from two different but similar receptors to build a new one could be a general approach to embrace during making biosensors that are based on a foreign receptor.
The Native E.coli Pathway
Downstream the Fusion Kinase
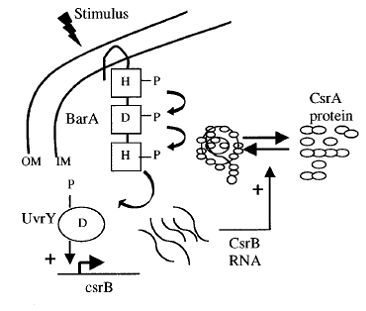
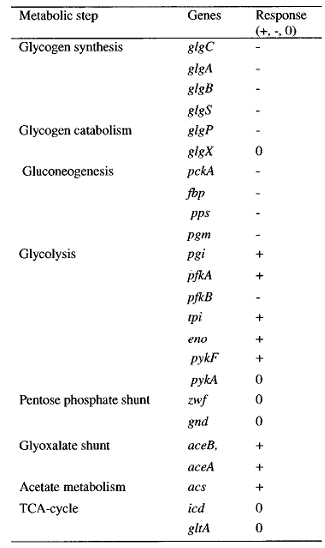
The sensing event triggers the phosphorilation of the CqsS-BarA and stimulates the transfer of phosphate group downstream the signaling cascade. Naturally, E.coli passes the phosphate group from BarA to its response regulator UvrY. This particular protein then binds to DNA triggering the transcription of small regulatory RNA called CsrB - Carbon Storage Regulator B. The most complicated part of the pathway is that CsrB inhibits CsrA. CsrA in turn inhibits a whole set of genes depicted in the table on the left ( plus represents activation, minus - inhibition).
One of the genes that CsrA inhibits is responsible for biofilm adhesion protein formation - PGA. This particular operon was our target for reporter molecule insertion. More details about pgaABCD operon can be found below.
Reference for the images: [2]
Fluorescent Reporter
Introduction
The initial plan was to introduce a Green Fluorescent reporter molecule under one of the genes controlled by small regulatory RNA called CsrA. So as not to reduce the viability of the cell as well as to avoid any possible “noise” in the engineered system, the gene choice required several attempts.
Re-usable testing kit
The plan to have a micro – chip for sensing biological water contamination ideally would include a possibility of reusing it. Thus, the aim of our project was to use GFP with an LVA tag (AANDENYALAA). This would mean that the GFP would have an amino acid tag targeted by housekeeping proteases and would be degraded within 45 minutes once there is no more stimulation for gene expression. This in turn would mean that the E.coli system sensing V.cholerae presence in water would stop fluorescing once the concentration of CAI-1 molecules decreased up to tolerable level. [2]
The idea to include degradable GFP in our project was taken from the Andersen, J.B et al. (1998) New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria. Applied and Environmental Microbiology.
pgaABCD operon control
This particular four - gene locus in E.coli is responsible for synthesis of the biofilm adhesion protein called PGA. As this is a virulence factor, i.e. non-essential for growth in supplemented media, a possible insertion of GFP and possible disruption the operon would not cause lethal effects to our cells.
As far as it is known, the pgaABCD genes are subject to post-translational inhibition by the small regulatory RNA CsrA as well as activation by the cation-responsive regulatory protein NhaR. The first fact is to our advantage. In theory, once the binding of the CAI-1 to fusion kinase occurred, the CsrA inhibition would be releaved and the GFP would be expressed . Also, according to research it has been proven that the csrA deletion mutant had an impressive 3000 fold increase compared to the wild type.
Possible “noise” in the system
As mentioned before, one of the factors to be taken into account prior choosing the gene within which the GFP would be inserted is the interference or “noise” in the system. This possibility might have caused background fluorescence or GFP expression independent from the sensing event of our interest.
The reasons underlying this “noise” are listed below:
- High salt concentrations (NhaR dependent)
- Alkaline pH (NhaR dependent)
- Ethanol ( NhaR dependent)
- Glucose presence in the media (Independent from both CsrA and NhaR)
The noise in the system appears only if the cells are grown at extreme conditions. However, if the growth media contains only 1% glucose, 1 % ethanol, 1 % NaCl the levels of the pgaABCD transcript should not be elevated by any other factors except CsrA. On the other hand, if additional NaCl or glucose are added to the medium it might cause significant increase in the pgaABCD mRNA appearance [2]
Target of insertion
As the pgaABCD is a four-gene locus another question that arose was the actual place within the operon where the insertion could take place. Another obstacle was that genes b, c and d overlap with each other.
One would theoretically suggest insertion under the promoter sequence, before the actual gene a . However, the very high inhibition with CsrA is actually due to 6 binding sites, one of which overlaps with gene a formyl-methionine codon.
Thus, the only possible way of inserting GFP, with minimal disruption of the whole operon, is at the end of gene a sequence just before the termination codon. This would give a high level of control by CsrA, as it is just after the CsrA regulatory binding sites, and would hopefully prevent extensive damage to genes b,c and d expression.
Cholera Autoinducers
Cholera autoinducer (CAI) is a signaling molecule. It is unusual from most pathogen autoinducers as it actually inhibits virulence factor production at high concentration. This is because of the nature of the disease V.Cholerae causes. The inhibition of virulence factors allows the bacteria to be flushed from the host body into the environment.
CAI is used naturally only by V.Cholerae. This makes it very useful in our bacterial sensor, because unlike other autoinducers, such as AI-2, which are used by many bacteria, this allows us to know bacteria species detected.
CAI is a short ketone with a hydroxyl group. The image below is the different CAI molecules (3 isomer pairs)
The Microchip Theory
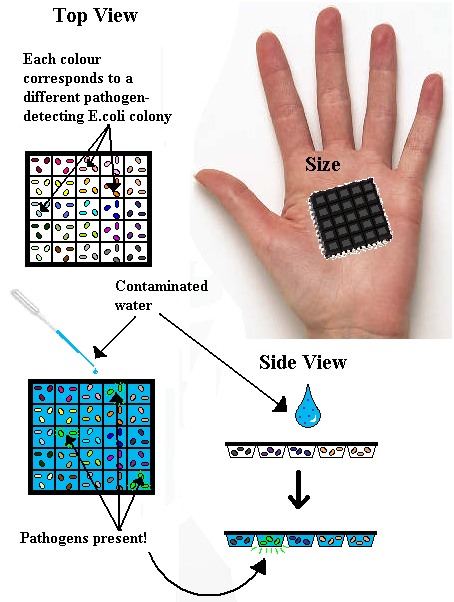
Our idea originated with the concept of a microchip that would test water for pathogen concentration. Whilst one summer wouldn’t allow us to actually build it, we still had an impression as to what it would look like.
How?
The target was for a cheap, reusable, quick and non-technical piece of equipment. At first, the microchip really was micro, and would require a microscope to look at, but in areas of natural disaster or extreme poverty this wasn’t viable. So we planned and hand-held size chip, with a number of ‘wells’ or ‘chambers’ built into it. Each chamber would contain a different pathogen-sensing E.coli colony. This presented other problems into how to keep the E.coli alive – could they survive if attached physically to the chamber? Or would they have to be frozen, or suspended in media? Frozen seemed the most likely, but not ideal, but proper design could be implemented later. Then the idea was to take a sample of water, possibly contaminated, and place it on the chip. The contaminants would interact with the specially designed E.coli, and those activated by contaminants would produce a distinctive glow visible to the naked eye. The chamber that glows corresponds to an unhealthy level of a particular pathogen in the water sample.
The reusable aspect came from the idea that disposable equipment is environmentally unfriendly. However, it could be cheaper than a reusable option, so if out plan were to be properly carried out, both chips should be developed to provide options. To make the chip reusable however, LVA tags are attached to the GFP in the DNA code, which attracts a housekeeping protease to cleave the GFP more rapidly than if left to degrade in the cell alone. A short burst of glow would be visible in cell, which would disappear by the time the chip dries out/the water passes through it.
Any takers?
This approach requires absolutely no technical knowledge, and could be ideal for its purposes. A future iGEM project anyone?
Parts and BioBricks
After various attempts to use the Biobrick booklet for transformation procedures needed for implied project ideas, we came to conclusion that the booklet shipped was actually faulty. Tests then proved this to be true. Thus the required BioBrick vector for submission of our designed parts was impossible to use from the booklet we were provided. Unfortunately, we ascertained this too late to receive variance approval. Which means we have BioBricks built but no way to send/enter them.
The BioBricks we made, however, were:
- 1.) CsrA, with promoter
- 2.) CsrA, without promoter
- 3.) CsrB, with promoter
- 4.) CsrB, without promoter
- 5.) UvrY, with promoter
- 6.) UvrY, without promoter
- 7.) CsqS
- 8.) CqsA
- 9.) BarA
- 10.) Red recombinase system!
- 11.) pKD46 Ori site - temperature sensitive
- 12.) FLP recombinase
Acknowledgements
We would like to thank to everyone who helped us on our way. That is to Prof. Visakan Kadirkamanathan whose excitement about our venture made us believe that it is possible. Prof. Philip Wright for providing us with a fine lab space in the Sheffield Bioincubator. Dr Catherine Biggs for supporting us with her knowledge and advice. Prof. David Hornby and his lab for being constant source of practical help. Esther Karunakaran was playing role of our advisor. She was a patient tutor that guided us through our project. Prof. Jeff Green and Prof David Rice for an advice regarding theoretical aspects of our project. Dr Graham Stafford, Bassler Lab and Ojar Melfors for providing us biological material for our experiments. All members of ChELSI group, especially Joy, for their constant advice on every day basis. Dave Wengraf who knows all legal aspect of science helped us with paper work and got us VAT exemptions. Prof. Robert Pool and Prof. Dominic Shellard for providing generous contribution toward our travel to iGEM Jamboree. IDT DNA for their sponsorship. IChemE, BioFusion and University of Sheffield Union of Students for supporting us financially. Without you this project would be much harder, if not impossible to carry out!
References
- 1.) Andersen, J.B et al. (1998) New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria. Applied and Environmental Microbiology
- 2.) Cerca, N. and Jefferson, K. K. (2008) Effect of Growth Conditions On Poly-N-acetylglucosamine expression and Biofilm Formation in Escherichia coli
- 3.) Goller, C. et al. ( 2006) The Cation-Responsive Protein NhaR of Excherichia coli Activates pgaABCD Transcription, Required for Production of the Biofilm Adhesin Poly-Beta-1, 6 - N - D - Glucosamine. Journal of Bacteriology
- 4.)Datsenko & Wanner, (2000), One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products
- 5.) Higgins, Bassler et al, (2007), The major Vibrio cholerae autoinducer and its role in virulence factor production
- 6.) Hammer & Bassler, (2007), Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae
- 7.) Jun Zhu, Melissa B. Miller, et al, (2001), Quorum-sensing regulators control virulence gene expression in Vibrio cholerae
- 8.)Tomenius, Pernestig et al, (2005), Genetic and functional characterization of the E.coli BarA-UvrY Two-componant system
- 9.) Suzuki et al, (2002), Regulatory Circuitry of thr CsrA/CrsB and BarA/UvrY systems of E.coli
- 10.) Sahu, Acharya et al, (2003), The bacterial adaptive response gene, barA, encodes a novel conserved istidine kinase regulatory switch for adaptation and modulation of metabolism in E.coli
- 11) Sabnis, A. N. et al., (1995), Pleiotropic Regulation of Central Carbohydrate Metabolism in Escherichia coli via the Gene csrA
- 12) Ytoh, J. et al., (2006), Roles of pgaABCD Genes in Synthesis, Modification, and Export of the Escherichia coli Biofilm Adhesin Poly-beta-1,6-N-Acetyl-D-Glucosamine
- 13) Yang., H. et al. , (1995), Coordinate Genetic Regulation of Glycogen Catabolism and Biosynthesis in scherichia coli via the CsrA Gene Product
- 14) Romeo, T. (1998), Global Regulation by the small RNA-binding protein CsrA and non-coding RNA molecule CsrB
- 15) Wang, X. et al. (2005), CsrA post-transcriptionally represses pgaABCD,responsible for synthesis of a iofilm polysaccharide adhesin of Escherichia coli
- 16) Romeo, T. et al. (1995), Identification and Molecular Characterization of csrA, a Pleiotropic Gene from Escherichia coli That Affects Glycogen Biosynthesis, Gluconeogenesis, Cell Size, and Surface Properties
- 17) Pernestig, A. K. et al. (2002), The Escherichia coli BarA-UvrY Two-Component System Is Needed for Efficient Switching between Glycolytic and Gluconeogenic Carbon Sources
- 18) Pernestig, A. K. et al. (2000), Identification of UvrY as the Cognate Response Regulator for the BarA Sensor Kinase in Escherichia coli
- 19) Mondragon, V. et al. (2006), pH-Dependent Activation of the BarA-UvrY Two-Component System in Escherichia coli
- 20) Rogov, V. V. et al. (2006), A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase onnects Histidine Kinase and Phosphoreceiver Domains
- 21) Tomenius, H. et al. ( 2006), The Escherichia coli BarA-UvrY two-component system is a virulence determinant in the urinary tract
- 22) Zhu, Y. and Inouye, M. (2004), The HAMP Linker in Histidine Kinase Dimeric Receptors Is Critical for Symmetric Transmembrane Signal Transduction
- 23) Zhu, y. and Inouye, M. (2008), Analysis of the Role of the EnvZ Linker Region in Signal Transduction Using a Chimeric Tar/EnvZ Receptor Protein, Tez1
- 24) Walters, M. and Sperandio, V. (2006), Quorum sensing in Escherichia coli and Salmonella
- 25) Gonzales, J. E. and Keshavan, N. D. (2006), Messing with Bacterial Quorum Sensing
- 26) Haseltine, E. L. and Arnold, F. H. (2007), Implications of Rewiring Bacterial Quorum Sensing
- 27) Alur, J. et al. (2002), Modeling and Analyzing Biomolecular Networks
- 28) Dunlap, P. V. (1999), Quorum Regulation of Luminescence in Vibrio fischeri
- 29) Sveningsen, S. L. et al. (2008), A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode
- 30) De Silva, R. S. et al. (2007), Crystal Structure of the Vibrio cholerae Quorum-Sensing Regulatory Protein HapR
- 31) Lenz, D. H. et al. (2005), CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae
- 32) Heidelberg, J. F. et al. (2000), DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae
- 33) Neidtich, M. B. et al. (2006), Ligand-Induced Asymmetry in Histidine Sensor Kinase Complex Regulates Quorum Sensing
- 34) Matson, J. S. et al. (2007), Regulatory Networks Controlling Vibrio cholerae Virulence Gene Expression
- 35) Hammer, B. K. and Bassler, B. L. (2003), Quorum sensing controls biofilm formation in Vibrio cholerae
- 36) Hammer, B. K. and Bassler, B. L. (2007), Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae
- 37) Lin, W. et al. (2005), Requirements for Vibrio cholerae HapR Binding and Transcriptional Repression at the hapR Promoter Are Distinct from Those at the aphA Promoter
- 38) Lenz, D. H. et al. (2004), The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio harveyi and Vibrio cholerae
- 40) Hoch, J. A. and Varughese, K. J. (2001), Keeping Signals Straight in Phosphorelay Signal Transduction
- 41) McLean, R. J. C. et al. (2004), A simple screening protocol for the identification of quorum signal antagonists
- 42) McClean, K. H. et al. (1997), Quorum sensing and Chromobacterium violaceum : exploitation of violacein
production and inhibition for the detection of N-acylhomoserine lactones
- 43) Martinko, J. M., Madigan, M. T. (2006) Brock Microbiology of Microorganisms. Pearson Benjamin Cummings, San Fransisco.
- 43) Vendeville, A. et al. (2005), Making‘SENSE’ Of Metabolism:Autoinducer!2, LuxS and Pathogenic Bacteria
- 44) Water, C. M. and Bassler, B. L. (2005), Quorum Sensing: Cell-to-Cell Communication in Bacteria
 "
"