Team:Michigan/Project/Fabrication
From 2008.igem.org
(Difference between revisions)
| Line 66: | Line 66: | ||
<br><br><font size=3 color=royalblue>NifA</font> | <br><br><font size=3 color=royalblue>NifA</font> | ||
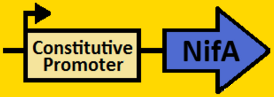
| - | *NifA was amplified from pRT22 using primers that had EcoRI, XbaI, NdeI and SpeI, PstI restriction sites flanking the 1560bp mutant sequence. | + | *NifA was [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol amplified] from pRT22 using primers that had EcoRI, XbaI, NdeI and SpeI, PstI restriction sites flanking the 1560bp mutant sequence. |
| - | *The NifA sequence was then purified using the QIAGEN QIAquick PCR Purification Kit. | + | *The NifA sequence was then [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol purified] using the QIAGEN QIAquick PCR Purification Kit. |
*NifA was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI and left overnight in 37⁰C. | *NifA was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI and left overnight in 37⁰C. | ||
*pArabLP-CP was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI also overnight at 37⁰C. | *pArabLP-CP was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI also overnight at 37⁰C. | ||
| Line 92: | Line 92: | ||
<font size=3 color=royalblue>NifHp</font> | <font size=3 color=royalblue>NifHp</font> | ||
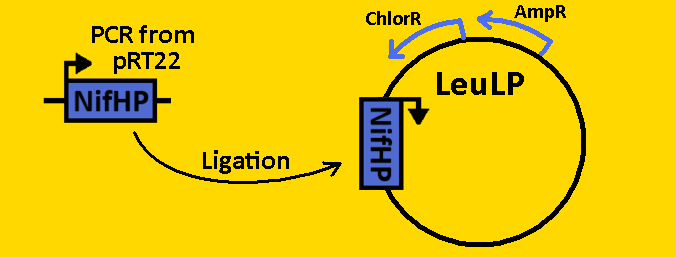
| - | *The NifH promoter (NifHp) was amplified from pRT22 using primers that had EcoRI, XbaI and NdeI, SpeI, PstI restriction sites flanking the 290bp sequence. | + | *The NifH promoter (NifHp) was [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol amplified] from pRT22 using primers that had EcoRI, XbaI and NdeI, SpeI, PstI restriction sites flanking the 290bp sequence. |
| - | *The NifHp sequence was then purified using the QIAGEN QIAquick PCR Purification Kit. | + | *The NifHp sequence was then [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol purified] using the QIAGEN QIAquick PCR Purification Kit. |
*NifHp was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI and left overnight in 37⁰C. | *NifHp was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI and left overnight in 37⁰C. | ||
*pLeuLP was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI also overnight at 37⁰C. | *pLeuLP was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI also overnight at 37⁰C. | ||
| Line 105: | Line 105: | ||
<br><br><font size=3 color=royalblue>NifL</font> | <br><br><font size=3 color=royalblue>NifL</font> | ||
| - | *NifL was amplified from pIM18 using primers that had EcoRI, XbaI, NdeI and SpeI, PstI restriction sites flanking the 1590bp mutant sequence. | + | *NifL was [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol amplified] from pIM18 using primers that had EcoRI, XbaI, NdeI and SpeI, PstI restriction sites flanking the 1590bp mutant sequence. |
| - | *The NifL sequence was then purified using the QIAGEN QIAquick PCR Purification Kit. | + | *The NifL sequence was then [https://2008.igem.org/Team:Michigan/Notebook/PCRProtocol purified] using the QIAGEN QIAquick PCR Purification Kit. |
*NifL was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI and left overnight in 37⁰C. | *NifL was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI and left overnight in 37⁰C. | ||
*pClock1 was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI also overnight at 37⁰C. | *pClock1 was double [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with NdeI and SpeI also overnight at 37⁰C. | ||
| Line 118: | Line 118: | ||
<br><br><font size=3 color=royalblue>GFP </font> | <br><br><font size=3 color=royalblue>GFP </font> | ||
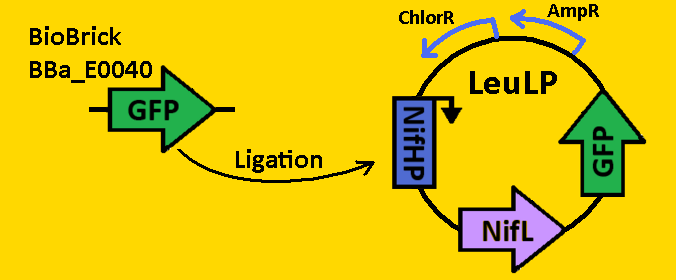
| - | *pClock2 was digested with EcoRI and SpeI | + | *pClock2 was https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI |
*BioBrick BBa_E0040 that contained GFP was [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and XbaI in sequential digests. | *BioBrick BBa_E0040 that contained GFP was [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and XbaI in sequential digests. | ||
*Digests were [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol purified] using QIAGEN Purification Kit | *Digests were [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol purified] using QIAGEN Purification Kit | ||
Revision as of 03:15, 30 October 2008
|
|---|
|
Project FabricationWe will be using landing pads to insert the Sequestillator onto the chromosome of E. coli. Noisy behavior has proven detrimental to clock studies throughout the past, and we hope to reduce the noise in our system using landing pads.
Landing Pad Plans for Sequestillator
Specific Fabrication TechniquesActivator Operon
Repressor Operon
|
|---|
 "
"