|
|
| Line 8: |
Line 8: |
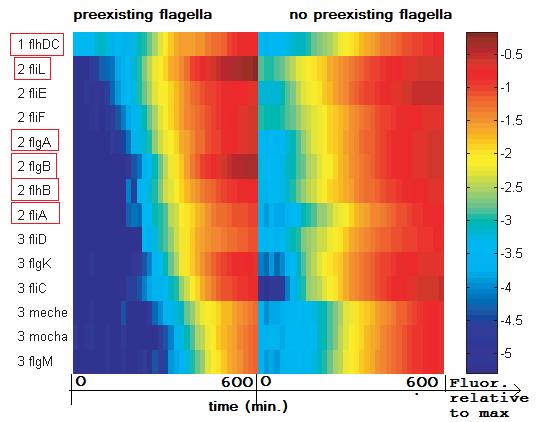
| | expression of the fl-genes observed in [[Team:Paris/Modeling/Bibliography#References|[1]]] | | expression of the fl-genes observed in [[Team:Paris/Modeling/Bibliography#References|[1]]] |
| | | | |
| - | == <FONT size=5> flhDC + fliA → pfliA → fliA → ∅ </FONT> <BR> == | + | == Oscillations == see [[Team:Paris/Modeling/Oscillations#Resulting_Equations|meanings of the constants]] |
| - | * is the contribution of flhDC and fliA on pfliA a SUM gate
| + | |
| - | [[Team:Paris/Modeling/Bibliography#flhDC_.2B_fliA_.E2.86.92_pfliL_.E2.86.92_Z1_.E2.86.92_.E2.88.85|(see fliL)]] ? | + | |
| - | * It seems that the FlhDC complex is an hexamere, as FlhD4-FlhC2.
| + | |
| | | | |
| - | == <FONT size=5> flhDC + fliA → pfliL → Z1 → ∅ </FONT> <BR> == | + | == FIFO == |
| | | | |
| - | | + | == Synchronisation == |
| - | the contributions of flhDC and fliA in the activity of pfliL appear to be a SUM
| + | |
| - | [[Team:Paris/Modeling/Bibliography#References|[2]]] : <br>
| + | |
| - | <FONT size=3> <center> act_pfliL(flhDC,fliA) = act_pfliL(flhDC,0)+act_pfliL(0,fliA) </center> </FONT> <BR> <br>
| + | |
| - | is it the same for the other promoters under the influence of both flhDC and fliA ?
| + | |
| - | | + | |
| - | == <FONT size=5> flhDC + fliA → pflgA → Z2 → ∅ </FONT> <BR> ==
| + | |
| - | is the contribution of flhDC and fliA on pflgA a SUM gate
| + | |
| - | [[Team:Paris/Modeling/Bibliography#flhDC_.2B_fliA_.E2.86.92_pfliL_.E2.86.92_Z1_.E2.86.92_.E2.88.85|(see fliL)]] ?
| + | |
| - | | + | |
| - | == <FONT size=5> flhDC + fliA → pflgB → Z2 → ∅ </FONT> <BR> ==
| + | |
| - | is the contribution of flhDC and fliA on pflgB a SUM gate
| + | |
| - | [[Team:Paris/Modeling/Bibliography#flhDC_.2B_fliA_.E2.86.92_pfliL_.E2.86.92_Z1_.E2.86.92_.E2.88.85|(see fliL)]] ?
| + | |
| - | | + | |
| - | == <FONT size=5> flhDC + fliA → pflhB → Z3 → ∅ </FONT> <BR> ==
| + | |
| - | is the contribution of flhDC and fliA on pflhB a SUM gate
| + | |
| - | [[Team:Paris/Modeling/Bibliography#flhDC_.2B_fliA_.E2.86.92_pfliL_.E2.86.92_Z1_.E2.86.92_.E2.88.85|(see fliL)]] ?
| + | |
| - | | + | |
| - | == <FONT size=5> lasI → 3OC12HSL → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> d[HSL]/dt = -D * Grad²([HSL]) </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> lasR → Rec → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> 3OC12HSL + Rec → HSL-Rec → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> HSL-Rec + pRep → TetR → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> ∅ → aTc → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> TetR + aTc → pTet → flhDC → ∅ </FONT> <BR> ==
| + | |
| - | | + | |
| - | == <FONT size=5> OmpR* → pflhDC → flhDC → ∅ </FONT> <BR> ==
| + | |
| | | | |
| | == <center> '''References''' </center> == | | == <center> '''References''' </center> == |
 "
"