Team:Hawaii/Ligation of pRL1383a Parts
From 2008.igem.org
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
Ligation of Parts
- The BioBrick parts of pRL1383a are to be ligated in a series of experiments.
Methods
Restriction Digest
- Each part is digested with both enzymes. For all cases NEBuffer 2 is used.
- Reaction Conditions:
- 5ul Buffer
- 1ul Enzyme 1 + 1ul Enzyme 2
- 0.5 ul BSA
- Xul water
- Yul insert (1ug if possible)
- Zul vector (less than 1ug)
- Running Conditions: 2 hours at 37°C.
- Check the progress of the reaction by running a 0.8% gel.
Ligation
- Refer to http://openwetware.org/wiki/DNA_ligation for the reasoning behind this.
Transformation
Verification with Colony PCR
- Reaction conditions: 10ul volume: 5ul green taq, 4ul water, 1ul primers (VF2+VR)+ a sterile poke from a colony.
- Running conditions: 94 hold, [95 30", 62 30", 72 (time varies based on size of verification site)]x 30 cycles, 72 10', 4 hold.
Results
7/27
- A few re-digests and ligations were performed today: oriT + pSB1A3, oriV + pSB1A3, mob + B0024, rep + R0010, aadA(pRL1383a+R0010). Afterwards, the ligation product was transformed to DH5-alpha. Only OriT transformed. I did not check the progress of this experiment with a gel, so I am not sure where this went wrong.
- Next time, a gel will be run after every step.
| Name | size | enzyme | quantity |
|---|---|---|---|
| oriT | ~125bp | XbaI & PstI | n/a |
8/11
| Name | size | enzyme | quantity |
|---|---|---|---|
| rep | 3.3kb | XbaI & PstI | 5ng/ul |
| oriV | 415bp | XbaI & PstI | 5ng/ul |
| aadA (pRL1383a) | 806bp | XbaI & PstI | 5ng/ul |
| aadA (BB) | 806bp | XbaI & PstI | 5ng/ul |
| P1 lytic Region | 1.3kb | EcoRI & SpeI | 5ng/ul |
| [http://partsregistry.org/Part:BBa_B0030 BBa_B0030] | 2094bp | SpeI & PstI | 4ng/ul |
| [http://partsregistry.org/Part:BBa_B0015 BBa_B0015] | 3318bp | EcoRI & XbaI | 5.7ng/ul |
| [http://partsregistry.org/Part:pSB1A2 pSB1A2] | 2094bp | SpeI & PstI | 5.5ng/ul |
| Name | strain, antibiotics | colonies? after next day??? | PCR verification in bp theoretical, experimental |
|---|---|---|---|
| rep(20ng)+B0030(16ng) | DH5-alpha & Amp100 | lawn | 3.3kb, 300&600 |
| oriV(20ng)+pSB1A2(50ng) | DH5-alpha & Amp100 | 21 colonies | 676, 600 |
| aadA (BB)+ B0030 | DH5-alpha & Amp100 | lawn | 900, #4:900, others: 300 |
| aadA(pRL1383a)+B0030 | DH5-alpha & Amp100 | lawn | 900, 300 |
| P1 lytic + B0015 | DH5-alpha & Kan50 | no colonies | n/a |
Transformation & Verification PCR
- The only bands corresponding to the correct size are the 4 oriVs and the 4th BB version of aadA.
- The rep region lanes are all either 300 or 600bp when they should be 3.3kb. If the RBS (B0030) ligated back to itself, the band should be 253bp. The bands at 300bp may be explained by this. It is unclear what happened in the latter case. The incorrect aadA bands are also around 300bp which would mean the RBS containing vector ligated back to itself some how. B0030 was cut with SpeI and PstI.
Plasmid Prep
- OriV1-4 and aadA (BB) were cultured over-night in Terrific Broth with amp100 as selection.
- The plasmids were isolated using an Alkaline Lysis Mini-Prep.
8/13
- Three ligations were performed today. I checked my math from the last ligation and decided it wasn't totally correct for the rep ligation to B0030.
- P1 lytic was ligated to B0015 (TT), Rep to B0030 with precisely 2:1 insert to vector ratio, and the aadA region from pRL1383a to B0030.
- Unfortunately there is no gel to confirm this ligation.
Transformation
- The ligation products were transformed into DH5-alpha cells with selection on LB amp100 plates. pUC18 was transformed into DH5-alpha cells as a positive control, while no plasmid was used in a transformation as a negative control.
- A lawn of colonies was observed for all three transformations. No colonies grew for either control (need to check pUC18 and see if there are problems with our stock).
PCR Verification
- The rep region, as with the last ligation was not inserted to the vector. The band at 300bp suggest the B0030 vector was amplified by VF2&VR. Another possibility is that some of this plasmid was not digested and the ligation just did not work, so these were the only circular plasmids available for transformation.
- The P1 lytic region is at 400kb while the verification size of B0015 is 445bp...
- aadA from pRL1383a is amplified to several different sizes. One of which is about correct (~1kb), one above 10kb, and one at 0.25kb.
9/2
9/6
- 3A-(ish) ligation is going to be used for subsequent ligation reactions.
- The lac promoter and RBS have been synthesized. They were ligated together 1:1.
- The other parts are to be digested out of plasmid or PCR amplified and restriction digested to give the correct over-hangs.
- Vector: E,P
- Front part: E,S
- Back: X,P
9/19/08
- Psuedo- 3A Assembly. This method is the same in that 2 pieces are assembled into a vector at one time, but differs in that the inserts are gel purified, so that the only plasmid involved is the recipient plasmid.
- Protocol:
- Restriction Digest: front insert (E,S), back insert (X,P), vector (E,P)
- Purification/Preparation of Parts: the vector is de-phosphorylated with SAP and the inserts are gel purified, with the exception of parts <200bp, which are ligated directly
- Ligation: at 16°C for 2 hours using T4 ligase.
Transformation of ligation Products
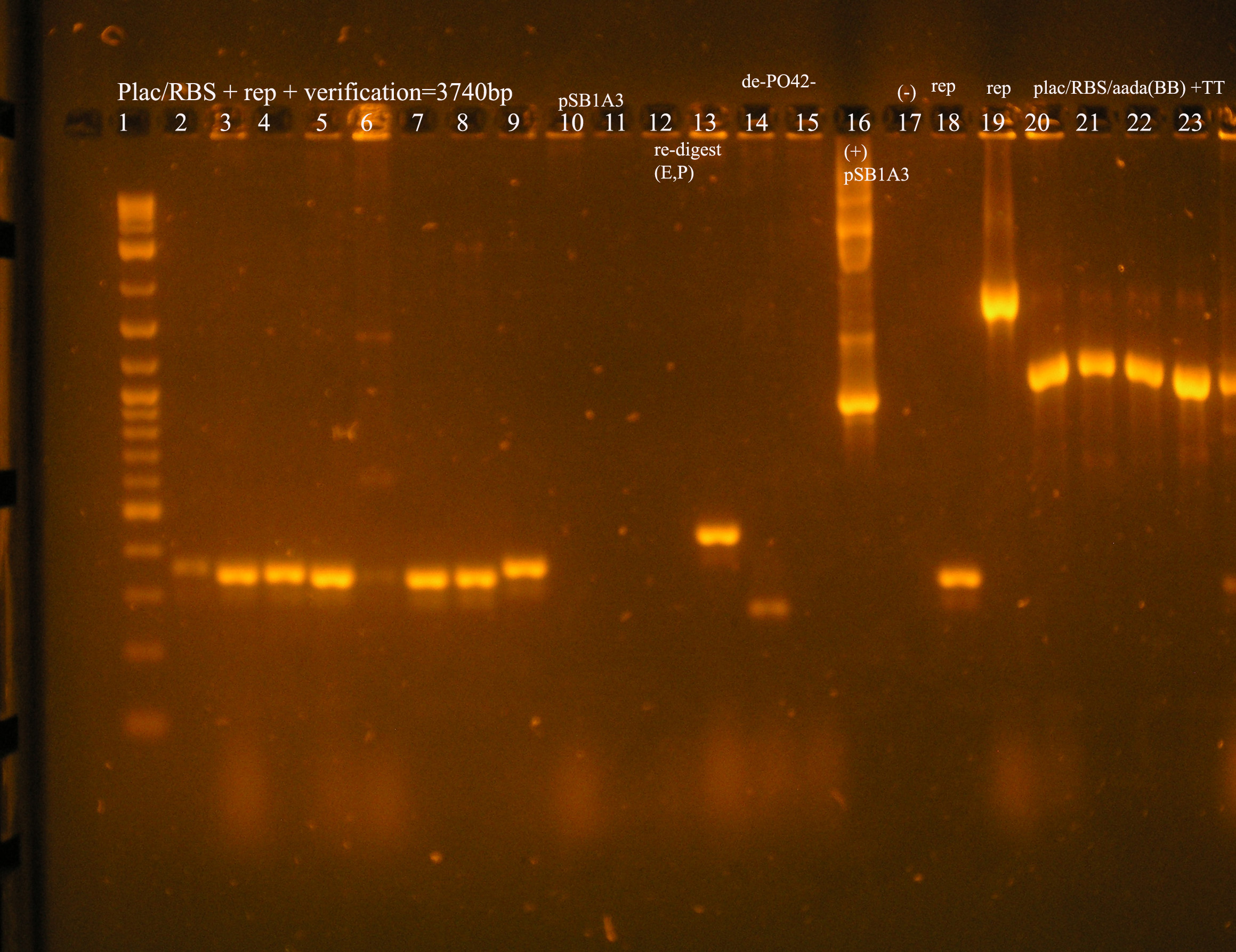
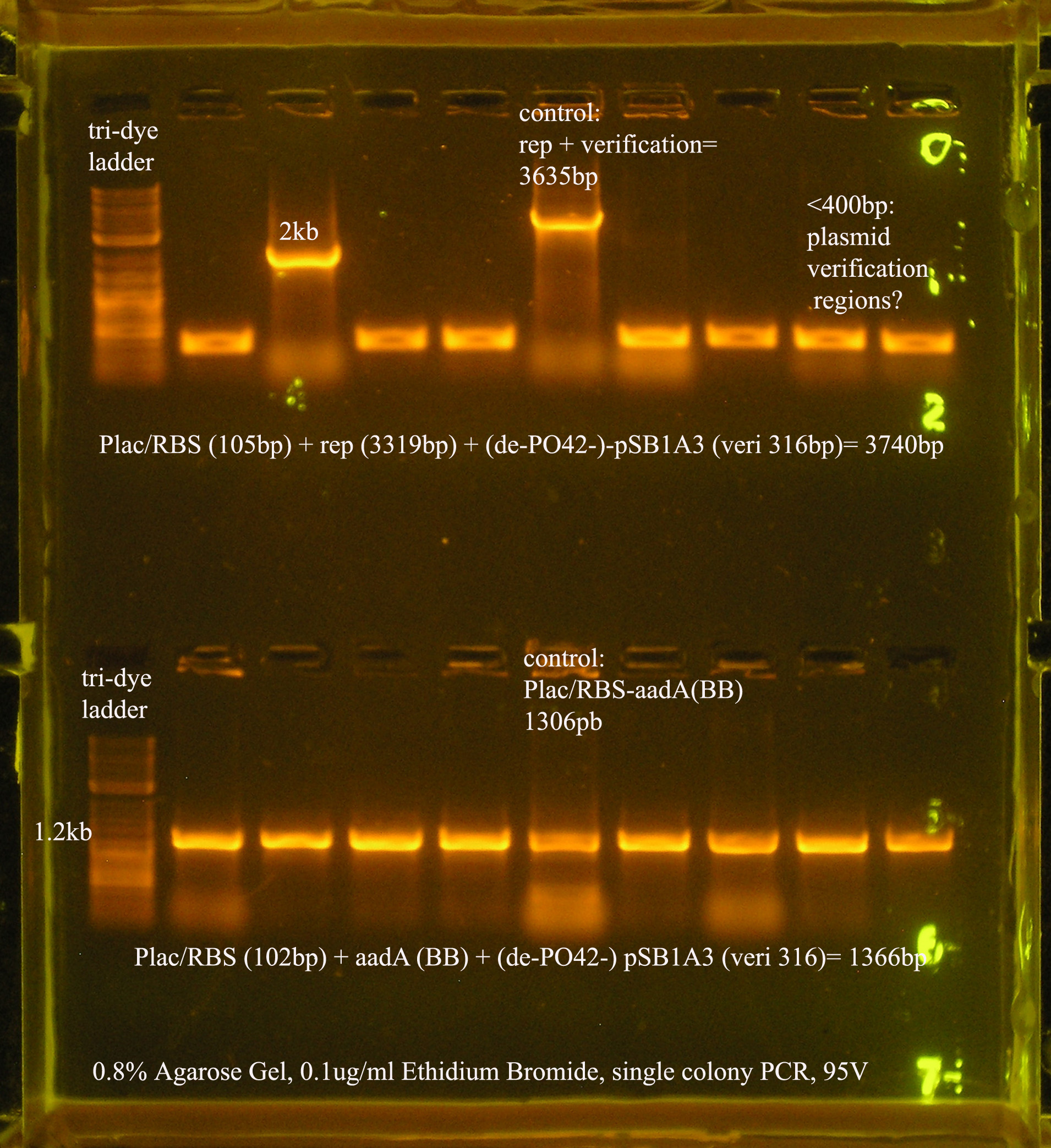
| Ligation Product/Plasmid, Ligated to de-phosphorylated pSB1A3 unless otherwise stated | Colonies | notes |
|---|---|---|
| Plac/RBS+rep | 71 | 16 colonies PCR'ed, compared to rep, none correct, PCR more |
| Plac/RBS/aadA(BB)+TT | 83 | 8 colonies PCR'ed, compared to Plac/RBS/aadA(BB), possibly larger than control, gel is warped, sequence |
| Plac/RBS/aadA(pRL1383a)+TT | 2 | 2 colonies PCR'ed, compared to Plac/RBS/aadA(pRL),larger than control, sequence, use |
| oriV | 11 | 11 colonies PCR'ed, 7 correct size (714bp), sequence |
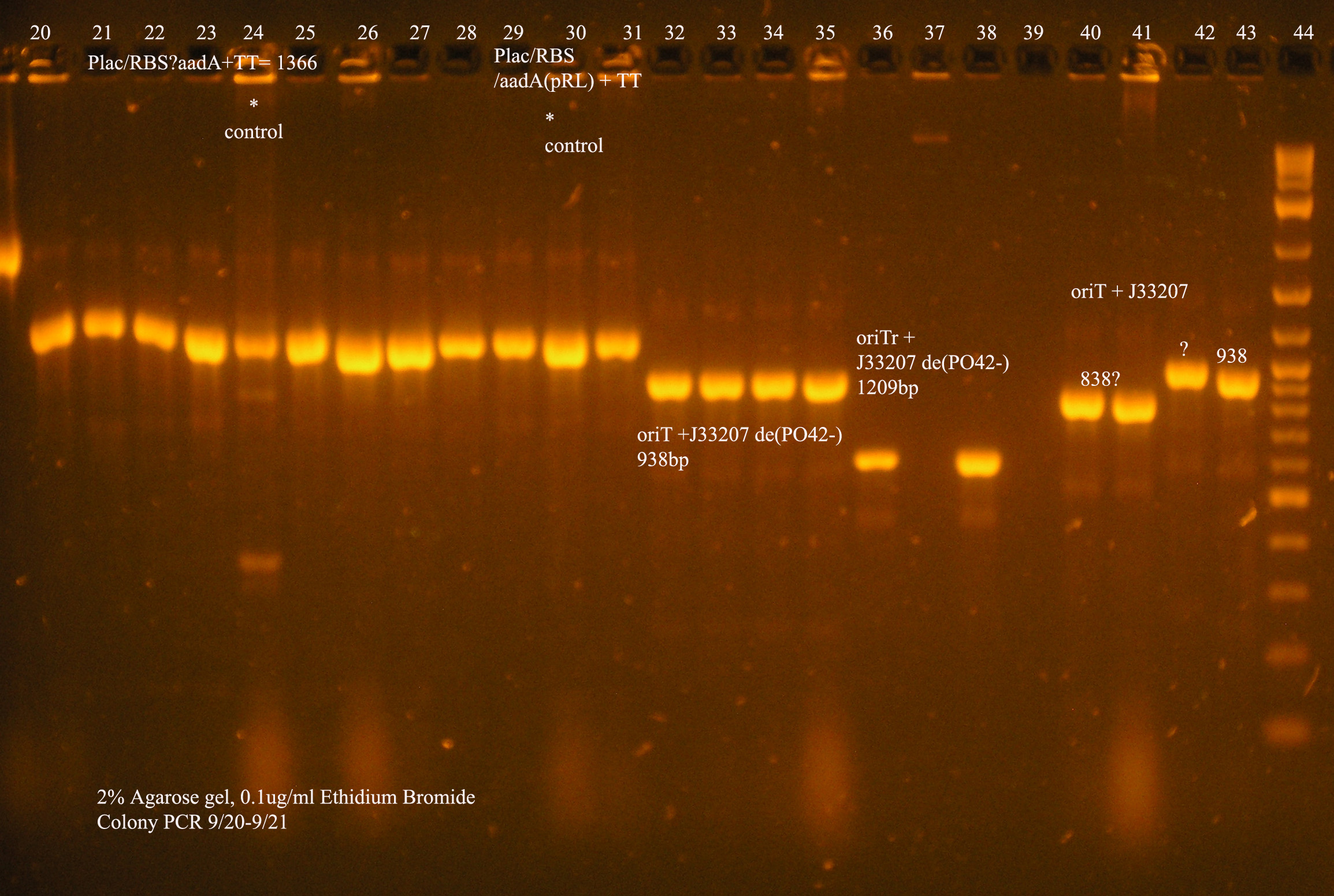
| oriT + J33207(de-phosphorylated) | 74 | 4 PCR'ed, all correct size, conjugation experiment, maybe sequence |
| oriTr + J33207(de-phosphorylated) | 89 | 4 PCR'ed, 0 correct (1209bp), 2 600bp |
| oriT + J33207 | 15 | 4PCR'ed, 1 correct, 2 size of J33207 insert, 1 ? |
| pSB1A3 | 7 | 2 PCR'ed, no amplification (VF2+VR) |
| pSB1A3 cut with (E,P) | 6 | 2 PCR'ed, 1= 450bp (VF2+VR) |
| pSB1A3 cut with (E,P), de-phosphorylated | 2 | 2 PCR'ed, 1= no amplification, 2= 250bp (VF2+VR) |
10/6
- Update on parts completed and those still in progress.
Individual Parts
- Plac/RBS/rep in pSB1A3
- PCR verification of this part was compared to rep and the size difference determined that further verification was needed.
This amplicon was sent in for sequencing using a series of primers for full coverage of sequence. rep was previously sequenced and contained BioBrick sites and no mutations, except for a 50 bp region at 1500 bp. An additional primer was designed to cover this region.
- A glycerol stock of this part was made for long-term storage.
- XbaI and PstI were used to digest Plac/RBS/rep in pSB1A3. This digest was run on a gel and purified.
- The next step is to ligate this part to oriV.
- Today a plasmid prep of the glycerol stock was prepared to ensure that I have this part in stock and that the re-digest can be replicated.
- Plac/RBS/aadA
- plac/rbs was added to aadA.
- a terminator was added to plac/rbs/aada. from the gel, it appear that something was added, however after sequencing these two constructs, it was found that the terminator was not added, however the BioBrick cloning sites, the promoter, rbs and aada all have correct sequences.
- a glycerol stock of plac/rbs/aada/terminator was prepared, though now I know there is no terminator in this construct. The final plasmid will have a transcription terminator near the aada region.
- plac/rbs/aada in pSB1A3 was cut with E,S and purified from a gel.
- oriV
- The most recent attempt to clone oriV into pSB1A3. From the gel, one would suspect that the ligation was successful, but the PCR verification of the 4 colonies yielded from the transformation were not amplified by VF2/VR.
- I will attempt the transformation again with the same ligation reaction.
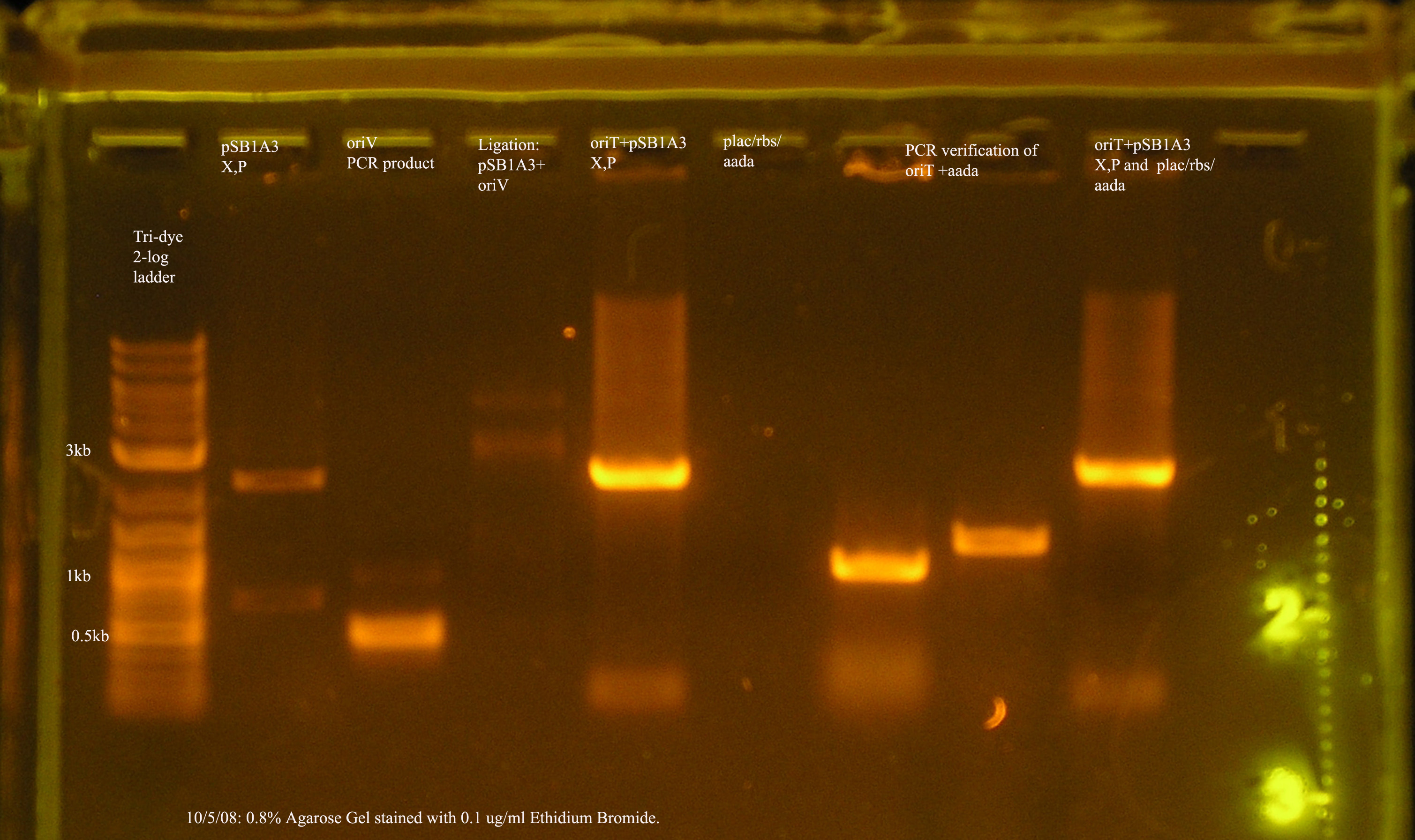
- oriT
- The sequence of this construct has been verified and oriT is stored on pSB1A2 in a glycerol stock.
Assembly of Parts
- oriT + plac/rbs/aada + pSB1A3
- The ligation and transformation were unsuccessful, and after running the two inserts and the pcr verification on a gel, it is clear that the plac/rbs/aada construct was not successfully purified from the gel. The band was visualized before purification, but not afterwards. I have started a culture from the glycerol stock of plac/rbs/aada.
- oriV + plac/rbs/rep + pSB1A3
10/20
- Sequencing of Ligation of [promoter/rbs/rep] came back and the promoter and rbs were not present. While I thought I had the construct [promoter/rbs/rep/oriV/promoter/rbs/aadA/oriT] in pSB1A3, I actually have [rep/oriV/promoter/rbs/aadA/oriT]. I will take the following steps:
- Front ligation of [promoter/rbs]into [rep/oriV/promoter/rbs/aadA/oriT]: cut [promoter/rbs] with E,S and cut [rep/oriV/promoter/rbs/aadA/oriT] with E and X and de-phosphorylate, then Ligate [promoter/rbs] with E,S with [rep/oriV/promoter/rbs/aadA/oriT]E,X,de-phosphorylated using T4 ligase. The negative control for this experiment is [rep/oriV/promoter/rbs/aadA/oriT]E,X,de-phosphorylated self-ligated.
- Results of Transformation:
Transformation of ligation Products
| Ligation Product | Colonies | notes |
|---|---|---|
| colony #13 [rep/oriV/promoter/rbs/aadA/oriT]E,X,de-phosphorylated + [promoter/rbs] | 80 colonies | colony pcr of 20 |
| colony #16 [rep/oriV/promoter/rbs/aadA/oriT]E,X,de-phosphorylated + [promoter/rbs] | 45 | colony pcr |
| colony #13 [rep/oriV/promoter/rbs/aadA/oriT]E,X,de-phosphorylated self-ligation | 11 | the de-phosphorylation was not complete? |
| pUC18 (+) control | lawn | |
| no plasmid (-) control | 0 |
- PCR was used to verify some of the colonies. I used the VF2/VR primers to identify if the correct insert is present. I also used some additional primers to identify whether or not the rbs was present:
Part Existence and Distance Verification
| name | primer | length | g/c | Tm | Reviewed By | Notes |
|---|---|---|---|---|---|---|
| VR_B0034 | gctACTAGTAtttctcctctttCTC | 25 bp | 40.0% | 56.3 C | NW | Use with VF2 primer to verify existence of, and approximate RBS B0034's distance from VF2 (may not work if something is post-fixed behind the RBS, since SpeI site could be destroyed (please double check)) |
- Gel results: The lane with (**) above it is an example of the expected results. In the top half of the gel, the colony was verified with VF2/VR primers. For this construct to be correct, we would expect a band at around 5.2 kb.
There is some non-specific binding in the lower kb region, but there is a band (*) that is around 5.2 kb. In the lane directly below this, the same colony was verified using the VF2/VR_B0034 primers. We would expect a band about 200 bp verifying the rbs located in front of the rep genes and a band at around 1 kb verifying that the aadA region has an rbs in front. This is verified by a positive control: the [promoter/rbs/aada] construct in pSB1A3 in a colony, pcr amplified with VF2/VR_B0034 primers. Both of these regions should be the same size because the amplification region should be the Forward verification region (158 bp) + the promoter (~60 bp) = 218 bp. The aadA regions was not amplified in this case, but due to the shortage of colonies containing results near what was expected, this colony, along with the colony designated with 3 * were cultured and plasmid prepped.
- The
Discussion
- Ligations into empty vectors seems to be successful, but when adding my part to an existing part, I run into problems.
- As a team we decided to synthesize the Promoters, RBS, and TT.
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"