|
__september
09-10-2008
CMV PCR
I did a PCR with primer for the CMV promotor. As DNA template I used the CMV+RLuc construct from the Ljubljana group which I isolated from the parts collection 2007.
For a 50 µl reaction I used:
40,4 µl H2O
5 µl buffer (10x)
1,5 µl FWD Primer (15pmol)(
1,5 µl REV Primer (15pmol)
1 µl dNTP (10mM)
0,1 µl DNA template
0,5 µl Pfu Polymerase
The settings for the PCR machine are the following:
1. T=94°C 00:02:00
2. T=94°C 00:00:30
3. T=62°C 00:00:30
4. T=72°C 00:01:00
5. GOTO 2 REP 29
6. T=72°C 00:10:00
7. HOLD 6°C
I got no product.
09-11-2008
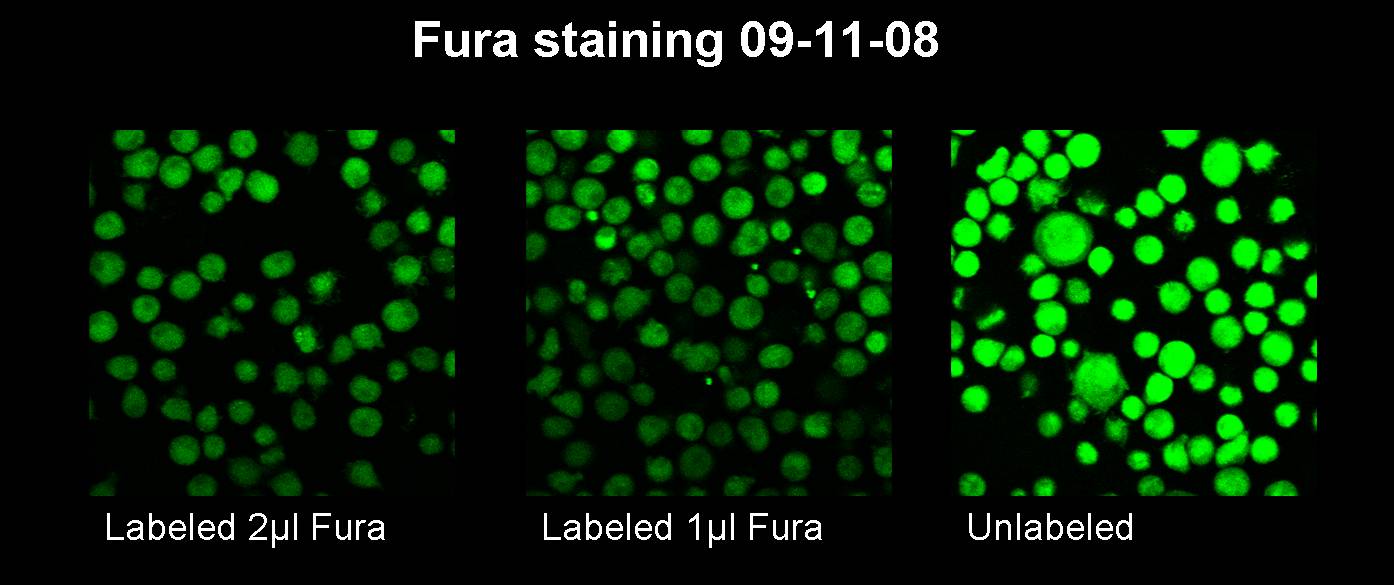
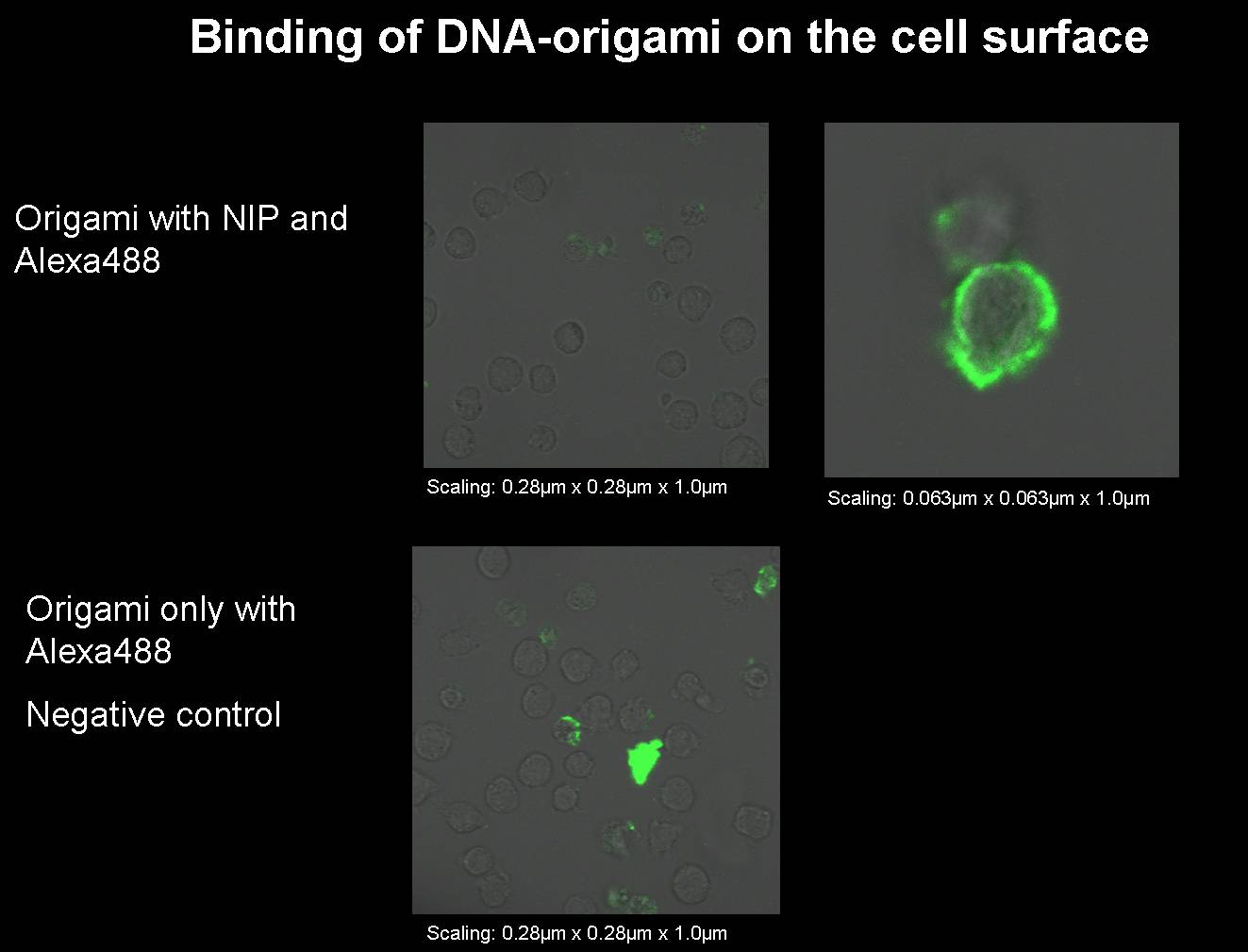
09-12-2008
1) Origami with NIP and
fluorophor for the
binding measurement
We
had to
produce some new origami for our next binding measurements.
- Origami with NIP and fluorophor
- Origami only with fluorophor
(without NIP);
negative control
see
at the
protocol from 07-24-2008
2)
Origami for the Calciummeasurement
- Origami without NIP (negative
control)
see at the
protocol from 07-24-2008
To increase the concentration
of
origami we also made to probes with the double amount
ingredients of the protocol
from 07-24-2008
|
|
Origami with NIP (6x (1:5)) [µl]
|
Origami without NIP (6x (1:5)) [µl]
|
|
Oligos-Pool
|
43,68
|
43,68
|
|
remainders
|
2,4
|
2,4
|
|
MgAc
|
1
|
1
|
|
Phage DNA (448,4 mg/µl)
|
33.6
|
33.6
|
|
NIP-Oligo
|
1,68
|
---------------------
|
|
Pool oligo without fluorophor
|
0,72
|
0,72
|
|
Oligo without NIP
|
-------------------
|
1,68
|
3) Master cycler
The origamis were produced in
the
mastercycler as explained before.
4) Purification of the DNA Origami
Was done as before
5) Digestion of CMV+Rluc
Digestion with EcoRV und FspI(3h at 37°C)
- 5µl plasmid
- 10µl H2O
- 0,5µl enzyme
- 2,5µl buffer (2)
- 0,5µl BSA
File:Verdau CMVRluc EcoRV FspI klein.jpg
6) CMV PCR
I tried the same PCR with different annealing temperatures. The optimal annealing temperature is 62°C. I used a gradient between 58°C and 62°C. I again got no products.
09-15-2008
CMV PCR
I again tried the CMV PCR, but this time with different polymerases. I used the Taq Polymerase and a Mix. I also tried one approach with the complete plasmid and one with the digested plasmid (see digestion of CMV+Rluc)
I got products in the approach with taq polymerase as well as in the approach with the Mix. I cut out 3 bands from each approach.
09-16-2008
Gel purification
I did a gel purification of the PCR products.
Digestion of the PCR products and the transfectionvector
Digestion with EcoRI and PstI. Then Kathrin did a ligation.
09-18-2008
Transformation
I did a transformation with the ligation product.
09-19-2008
Transformation
There were no colonies on the plates.
09-20-2008
Digestion of the PCR products and the transfectionvector
Now I used the restrictionenzymes XbaI and SpeI.
Gel purification and ligation
The digested PCR products and the vector.
09-21-2008
Transformation of the ligation
I used 10 µl of the ligation and used RV 308 cells
|  "
"


