Team:ETH Zurich/Wetlab/Genome Reduction
From 2008.igem.org
|
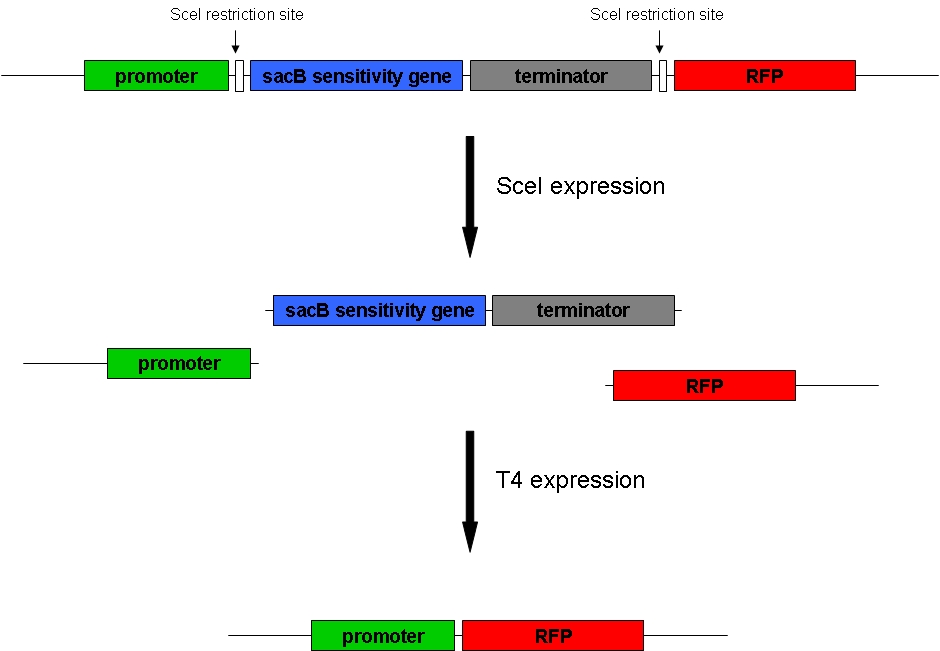
Genome reduction1. GoalGoal of our project is to randomly delete chromosomal fragments of E. coli in order to reduce the physical amount of genomic DNA. 2. MethodTo reach our goal of random deletion of chromosomal fragments, we want to express a frequently cutting restriction enzyme along with the simultaneous, shortly delayed, or even continuous expression of a ligase. The restriction enzyme will cut genomic DNA in a random fashion in vivo, while the ligase performs its job of religation. Assuming that the genomic DNA is cut at several sites within one cell, religation will lead to the exclusion of chromosomal fragments in some cells. Multiple rounds of restriction and religation will therefore lead to a markedly reduced genome. 3. Proof of conceptIn 1997 Ren et al. showed that in vivo religation of linearized vector and insert is possible by overexpression of the T4 ligase. Our idea, on the other hand, relies on the assumption that in vivo restriction and religation is possible and might lead to the exclusion of chromosomal DNA. Proving these assumptions is addressed in separate experiments and will from now on be called our “proof of concept”. Our proof of concept relies the following construct (not shown are ribosomal binding sites behind both SceI restriction sites and a terminator following the RFP): This construct was ordered at GeneArt and subsequently supposed to be integrated into the genome of a wild-type E. coli strain (MG1655). We are planning to use the lambda red recombination system established by Wanner and Datsenko (xxx) to integrate our construct into the tryptophanase A gene resulting in the knockout of the latter. Transformation of a plasmid-encoded T4 ligase and SceI restriction enzyme, both of which were also ordered at GeneArt, into cells carrying the above construct was supposed to yield bacteria that could be induced to express T4 and SceI. Before induction of these enzymes, the cells do not express RFP. After induction of the restriction enzyme, SceI would cut the bacterial chromosome at the sites indicated above. The ligase would religate the construct, leading to the exclusion of the sensitivity gene and the terminator at least in some cells. These cells would finally express RFP and could therefore easily be identified: Additionally, adding sucrose to the medium would kill those cells that have not eliminated the sacB sensitivity gene. Therefore, only RFP-expressing bacteria would survive, which would presumably facilitate detection by several orders of magnitude.
3.1 Modification of proof of conceptUnfortunately, up to now we have not received our construct for the proof of concept. Therefore, we are working on a modified construct:
3.2 SceI restriction enzymeSceI is a site-specific homing endonuclease. It is extremely rare-cutting as it recognizes an 18 bp sequence. These properties make SceI a perfect restriction enzyme for our proof of concept where it should only cut at the sites on our constructs indicated above. However, for our goal of minimizing E. coli’s genome we will need to use a frequently cutting restriction enzyme which will potentially lead to severe damage of the cell’s genome. In order to limit this damage, we want to be able to pulse the expression of the restriction enzyme. Therefore, we are cloning an arabinose-inducible promoter (I0500) also in front of SceI. 3.3 T4 ligaseT4 is the commonly used ligase for in vitro cloning. For in vivo cloning, high levels of T4 are advantageous not only for improving the efficiency of religation leading to the exclusion of chromosomal fragments, but also for limiting DNA damage. However, constitutive overexpression might lead to immediate religation without the exclusion of chromosomal fragments. Therefore, we are cloning T4 behind an IPTG-inducible promoter (R0010), and several constitutive promoter of differing strengths. An alternative idea would be to clone both SceI and T4 behind the same promoter, so that induction would lead to simultaneous expression of both enzymes.
|
 "
"