Modeling
From 2008.igem.org

|
| Home | The Team | The Project | Notebook |
|---|
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
Contents |
Overview
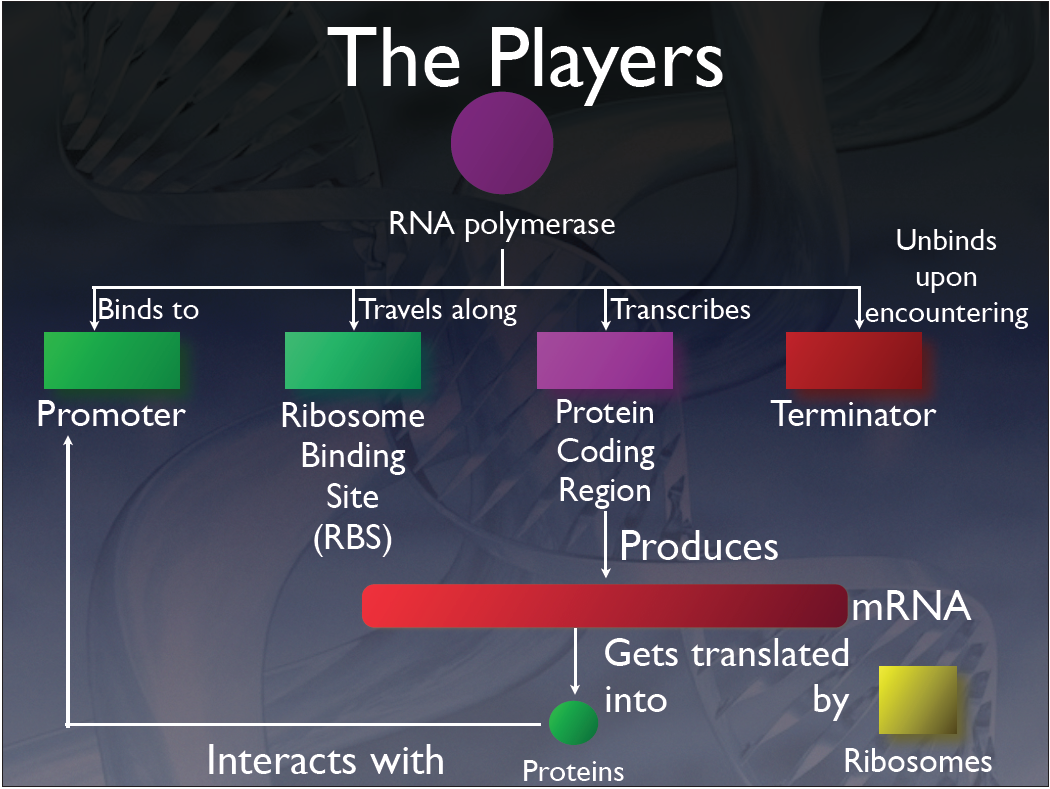
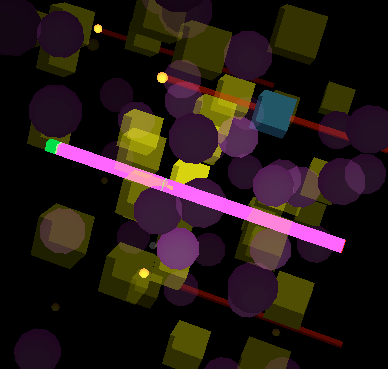
This schematic shows the agents involved in our simulation:
Transcription
The mRNA agents perform all of the functions that mRNA do in living cells, and it is directly related to the processes of transcription and translation. Once an RNA polymerase, which is represented visually as a large violet sphere, binds onto the DNA, the process of transcription occurs creating an mRNA. Upon the start of transcription, the RNA polymerase turns red, signifying that the process has started. As the process progresses, the mRNA that is being created gets gradually larger. When transcription finishes, the DNA releases the mRNA; it returns to its original violet color, and moves randomly within the space of the model. Due to the simulation software constraints, we could not represent mRNA as the thin ribbon-like shape that is seen in textbooks. Instead, we represented the mRNA visually as a long, thin, red rod. Contained within each mRNA is information required for the sequencing of proteins.
Translation
Once the mRNA is created, the ribosomes bind to the mRNA and perform the process of translation. Ribosomes in this simulation are represented as large yellow cubes. In reality, ribosomes are composed of two parts that attach to a mRNA on different sides upon translation. However, for the purposes of this simulation, we represented the ribosome with a single part. This prevents unnecessary complications and simplifies the computation. Since ribosomes are quite dense in cells, they are modeled with a relatively heavy mass in the simulation; their velocities and accelerations, as a result, are very small. When a ribosome attaches to an mRNA, the cubic ribosome turns into a teal colour to signify that translation is occurring. Since the mRNA is now attached to a heavier ribosome, the mRNA slows itself down to move at the same speed as the ribosome. When attached, the ribosome continues with translation by moving along the mRNA. Upon completion, protein molecules are released into the cell. In some cases, the mRNA codes for multiple protein molecules. As a result, some of the proteins are released during translation instead of at the end. Regardless, after translation has finished the ribosome detaches itself, once again becoming yellow and free to float within the simulation space.
mRBS
We created another class, the mRBS (messenger ribosomal binding site), which links the ribosome and mRNA classes; it is the hub for communication. Functionally, this class allows for the process of translation to occur; in addition, it presents the possibility of transcription and translation to occur nearly simultaneously, which is what usually occurs in prokaryotic cells. Visually, the mRBS is represented as a small orange sphere that is attached to the front of a mRNA. This mRBS is the site where translation occurs.
Half-Life
To prevent cluttering in the simulation - and to better mimic the reality of prokaryotic cells - we incorporated half-life. Half-life allows decay in molecules and proteins. mRNA and other protein molecules within the cell degrade or disappear in the simulation. However, components such as the RNA polymerase and the ribosomes are not subject to half-life
because we assume that they have a constant supply, which is the case in living cells.
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
| Home | The Team | The Project | Notebook |
|---|
 "
"