Team:Beijing Normal/Project
From 2008.igem.org
(→Special protocol) |
(→Special protocol) |
||
| (55 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {| style="color:# | + | {| style="color:#ffffff;background-color:#6699cc;" cellpadding="3" cellspacing="1" border="0" bordercolor="#fff" align="center" width="80%" |
| - | + | ||
!align="center"|[[Team:Beijing_Normal|Home]] | !align="center"|[[Team:Beijing_Normal|Home]] | ||
!align="center"|[[Team:Beijing_Normal/Team|The Team]] | !align="center"|[[Team:Beijing_Normal/Team|The Team]] | ||
!align="center"|[[Team:Beijing_Normal/Project|The Project]] | !align="center"|[[Team:Beijing_Normal/Project|The Project]] | ||
!align="center"|[[Team:Beijing_Normal/Parts|Parts Submitted to the Registry]] | !align="center"|[[Team:Beijing_Normal/Parts|Parts Submitted to the Registry]] | ||
| - | |||
!align="center"|[[Team:Beijing_Normal/Notebook|Notebook]] | !align="center"|[[Team:Beijing_Normal/Notebook|Notebook]] | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
== '''Overall''' == | == '''Overall''' == | ||
| Line 34: | Line 29: | ||
Taking the condition of our lab into account, we decide just deal with the aerobic degradation path way. And do some work on increasing the degradation effeciency. | Taking the condition of our lab into account, we decide just deal with the aerobic degradation path way. And do some work on increasing the degradation effeciency. | ||
| - | |||
| - | |||
== Project Details== | == Project Details== | ||
| Line 49: | Line 42: | ||
*Catabolic pathway for degradation of biphenyl and organization of bph gene cluster is as follows: | *Catabolic pathway for degradation of biphenyl and organization of bph gene cluster is as follows: | ||
| - | [[Image:PCBs pathway.jpg | + | [[Image:PCBs pathway.jpg|900px|center|thumb|Source: Kensuke Furukawa and Hidehiko Fujihara, '''Microbial Degradation of Polychlorinated Biphenyls: Biochemical and Molecular Features''', ''Journal of bioscience and bioengineering'', 105:433–449 (2008), permitted to use by Kensuke Furukawa]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | permitted to use | + | |
'''Enzymes responsible for oxidative degradation of PCBs''' | '''Enzymes responsible for oxidative degradation of PCBs''' | ||
| Line 75: | Line 64: | ||
In the forth step BphD, a hydrolase, hydrolyzes the ring meta-cleavage yellow compound(HOPDA) to chlorobenzoic acid and 2-hydroxypenta-2,4-dienoate. | In the forth step BphD, a hydrolase, hydrolyzes the ring meta-cleavage yellow compound(HOPDA) to chlorobenzoic acid and 2-hydroxypenta-2,4-dienoate. | ||
| - | === | + | === Design Abstract === |
Polychlorinated biphenyls (PCBs) are a group of organic pollutants that are persistent when released into the environment. Our task is to design an effective as well as intelligent PCBs degrader. Thus our work could be divided into two parts. One is to develop a sensing system for the detection of PCBs and activation of the downstream degradation pathway; the other is to enhance the degradation efficiency. | Polychlorinated biphenyls (PCBs) are a group of organic pollutants that are persistent when released into the environment. Our task is to design an effective as well as intelligent PCBs degrader. Thus our work could be divided into two parts. One is to develop a sensing system for the detection of PCBs and activation of the downstream degradation pathway; the other is to enhance the degradation efficiency. | ||
| Line 83: | Line 72: | ||
As dihydrodiols and dihydroxybiphenyls are very toxic to bacterial even after short incubation time, we design a feedback repression pathway use sRNA components— sodB and rhyB. Part of sodB was fusion with bphA and biosurfactant pathway and will be repressed by rhyB which is under the control the promoter Ppcb. As only when rhyB molecules is much more than sodB fusion mRNA, it will play a role of repression, only the high concentration of DHBP will arise this reaction. In this case, the amount of these two metabolites will be reduced in two ways. | As dihydrodiols and dihydroxybiphenyls are very toxic to bacterial even after short incubation time, we design a feedback repression pathway use sRNA components— sodB and rhyB. Part of sodB was fusion with bphA and biosurfactant pathway and will be repressed by rhyB which is under the control the promoter Ppcb. As only when rhyB molecules is much more than sodB fusion mRNA, it will play a role of repression, only the high concentration of DHBP will arise this reaction. In this case, the amount of these two metabolites will be reduced in two ways. | ||
| - | As to the sensor part, dihydroxylated PCBs are substrate of the clcA-encoded chlorocatechol dioxygenase and thus induce the clcR and related promoter, so we use this as the sensing system. And T7 amplification system is add downstream to amplify the signal. | + | As to the sensor part, dihydroxylated PCBs are substrate of the clcA-encoded chlorocatechol dioxygenase and thus induce the clcR and related promoter, so we use this as the sensing system. And T7 amplification system is add downstream to amplify the signal. |
| - | + | ||
=== The Experiments === | === The Experiments === | ||
| Line 125: | Line 113: | ||
To obtain some genes, we often have to use bacteria chromosome or large size plasmid as template in which high GC% content and complex secondary structures are seriously hampering PCR and thus leading to complete failure. To solve this problem, we have developed a special protocol-- hot start method combined with additive(sole or mixed)-- which is most helpful . | To obtain some genes, we often have to use bacteria chromosome or large size plasmid as template in which high GC% content and complex secondary structures are seriously hampering PCR and thus leading to complete failure. To solve this problem, we have developed a special protocol-- hot start method combined with additive(sole or mixed)-- which is most helpful . | ||
| - | + | === Simplified Hot Start PCR === | |
Hot start method is to prevent primer dimer and low specified product yield. In our experiment, We use common taq polymerase to replace commercial high-priced hot-start polymerase. | Hot start method is to prevent primer dimer and low specified product yield. In our experiment, We use common taq polymerase to replace commercial high-priced hot-start polymerase. | ||
After a 5min 95℃ pre-heat step, DNA polymerase is added to each PCR tube before the PCR cycling begin. After that, the regular PCR cycling could begin. | After a 5min 95℃ pre-heat step, DNA polymerase is added to each PCR tube before the PCR cycling begin. After that, the regular PCR cycling could begin. | ||
| - | + | ==== Explanation ==== | |
| - | + | ||
When reaction components are mixed at room temperature, reaction set up below the optimal primer annealing temperature, which permits nonspecific primer annealing and extention.Undesired, non-specific primer extetion products formed this way may be amplified in the PCR, resulting in misprimed products and primer ologomers.In hot start PCR, DNA polymerase is withheld from the mixture until the system has reached a temperature that favours specific primer annealing. As a result hot start PCR can greatly improve specificity, sensitivity and yield in a PCR. | When reaction components are mixed at room temperature, reaction set up below the optimal primer annealing temperature, which permits nonspecific primer annealing and extention.Undesired, non-specific primer extetion products formed this way may be amplified in the PCR, resulting in misprimed products and primer ologomers.In hot start PCR, DNA polymerase is withheld from the mixture until the system has reached a temperature that favours specific primer annealing. As a result hot start PCR can greatly improve specificity, sensitivity and yield in a PCR. | ||
| - | + | === Effective Additives === | |
| - | + | ||
The most simple additive in our PCR is DMSO(>=99.9% purity) at a concentration of 5%(V/V). It works well in most 'problematic' PCR, however, when DMSO fails in some case, we have to turn to a magic mixed additive (2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA) for help. This excellent additive has solved several PCR where even 5% DMSO fails. | The most simple additive in our PCR is DMSO(>=99.9% purity) at a concentration of 5%(V/V). It works well in most 'problematic' PCR, however, when DMSO fails in some case, we have to turn to a magic mixed additive (2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA) for help. This excellent additive has solved several PCR where even 5% DMSO fails. | ||
| Line 142: | Line 128: | ||
In certain PCR there is no yield without this additives, for example the ones by which we amplify bphA1A2A3A4 and bphBC. | In certain PCR there is no yield without this additives, for example the ones by which we amplify bphA1A2A3A4 and bphBC. | ||
| - | + | ==== Explanation ==== | |
One major factor limiting the output of PCR routines is that a number of DNA sequences are poorly or not amplifiable under standard reaction conditions, either because of their high GC-content or/and their instrict propertoties to form secondary structures. This protocol is intended for GC-rich DNA sequences. This is a concentration dependent combination of betaine, dithiothreitol, and dimethyl suloxide. According to the references the concentration ratio is: a mixture containing 2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA. It is stable at -20℃ for at least 3 months. | One major factor limiting the output of PCR routines is that a number of DNA sequences are poorly or not amplifiable under standard reaction conditions, either because of their high GC-content or/and their instrict propertoties to form secondary structures. This protocol is intended for GC-rich DNA sequences. This is a concentration dependent combination of betaine, dithiothreitol, and dimethyl suloxide. According to the references the concentration ratio is: a mixture containing 2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA. It is stable at -20℃ for at least 3 months. | ||
| + | === References for special protocol: === | ||
| + | 1. David E. Birch et al., '''Simplified hot start PCR''', ''Nature'' 381:445-446 (1996) | ||
| - | + | 2. S.Kaijalainen et al., '''An alternative hot start technique for PCR in small volumes using beads of wax-embedded reaction components dried in trehalose''', ''Nucleic Acids Research'' 21:2959-2960 (1993) | |
| - | + | ||
| - | + | 3. Yukihiro Kitade et al., '''Effect of DMSO on PCR of Porphyra yezoensis(Rhodophyta) gene''', ''Journal of Applied Phycology'' 15:555-557 (2003) | |
| - | + | 4. Markus Ralser et al., '''An efficient and economic enhancer mix for PCR''', ''Biochemical and Biophysical Research Communications'' 347:747–751 (2006) | |
| - | + | == Design Details == | |
| + | |||
| + | [[Image:p1.jpg]] | ||
| - | + | Enhance the degradation efficiency & detect PCBs | |
| + | |||
| + | ===1. Switch on the degradation pathway=== | ||
| + | |||
| + | [[Image:awake.gif]] | ||
| + | |||
| + | The system is awakened by PCBs. | ||
| + | |||
| + | |||
| + | We add bphR1 upstream the pathway and bphR2 downstream. bphR2 could sense the presence of PCBs and bphR1 could sense HOPDA, the third step product. These two regulators could switch on the degradation pathway. | ||
| + | |||
| + | In the absence of PCBs, small amounts of BphR2 protein bind to the bphR2 operator to re press bphR2 transcription (auto-repression). Under this condition, the levels of transcription of bph genes are very low. In the presence of PCBs, the BphR2 protein binds to the operators of bphR1 and bphA1A2A3A4BC and activates their transcription; thus high levels of HOPDA (the ring meta-cleavage compound) are produced from PCBs. In the presence of HOPDA, the transcription of bphR1 is further promoted, and its product BphR1 activates the transcription of bphR1 itself and downstream pathway [1,2]. | ||
| + | |||
| + | [[Image:t1.jpg]] | ||
| + | |||
| + | |||
| + | ===2. Solve the bottleneck in PCBs degradation pathway=== | ||
| + | |||
| + | Our idea: Add a spur track to solve the bottleneck of the pump. | ||
| + | |||
| + | [[Image:pump.gif]] | ||
| + | |||
| + | BphC (2,3-dihydroxybiphenyl 1,2-dioxygenase), the third enzyme of the bph pathway, is of particular significance because it is incapable o f catalyzing the ring cleavage step and is subject to various types of inhabitation, as well as suicide inactivation. According to the recent research, ortho-chlorinated PCB metabolites (DHBs) are potent and physiologically significant inhibitors of BphC, so we design a feedback activation pathway using small RNA to increase the BphC transcription and expression under a 2, 3-DHBP and 4-CB inducible promoter Ppcb [3]. As 2, 3-DHBP is the second metabolite in the pathway, 2, 3-DHBP at a concentration more than 0.1mM will induce the activate PpcbC and thus increase gene expression downstream. The activated threshold of PpcbC is relatively high, so this pathway will be activated when main PCBs degradation above has began. | ||
| + | |||
| + | [[Image:t2.jpg]] | ||
| + | |||
| + | ===3. Control on the amount of PCBs molecular that enter the cell.=== | ||
| + | |||
| + | As dihydrodiols (first step product) and dihydroxybiphenyls (DHBP, second step product) are very toxic to bacterial even after short incubation time, we design a feedback repression pathway use sRNA components— sodB and rhyB. Part of sodB was fusion with bphA and biosurfactant pathway and will be repressed by rhyB[4] which is under the control the promoter Ppcb. As only when rhyB molecules is much more than sodB fusion mRNA, it will play a role of repression, only the high concentration of DHBP will arise this reaction. This design will control the amount of PCBs molecular that enter through the membrane. | ||
| + | |||
| + | [[Image:t3.jpg]] | ||
| + | |||
| + | ===4. System guarantee=== | ||
| + | |||
| + | In our design T7 amplification system is used to amplify the signal. T7 polymerase is added downstream the main degradation pathway. When T7 polymerase is translated, it will activate the T7 promoter and downstream reporter gene, YFP. Then the signal could be detected. This system could be used to test the function of the degradation pathway. | ||
| + | |||
| + | [[Image:t4.jpg]] | ||
| + | |||
| + | |||
| + | ===References for Design Details:=== | ||
| + | |||
| + | [1] Kensuke Furukawa, et al., '''Microbial Degradation of Polychlorinated Biphenyls: Biochemical and Molecular Features''', ''Journal of Bioscience and Bioengineering'' 105:433–449 (2008) | ||
| + | |||
| + | [2] Stephen Y. K. Seah, et al., '''Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls''', ''Journal of Bacteriology'' 183:1511–1516 (2001) | ||
| + | |||
| + | [3] Sang-ho Park, et al., '''Construction of transformant reporters carrying fused genes using pcbC promoter of Pseudomonas sp DJ-12 for detection of aromatic pollutants''', ''Environmental Monitoring and Assessment'' 92:241–251 (2004) | ||
| + | |||
| + | [4] Qinhong Wang, et al., '''Engineering Bacteria for Production of Rhamnolipid as an Agent for Enhanced Oil Recovery''', ''Biotechnology and Bioengineering'', 98:842-853 (2007) | ||
| + | |||
| + | [5] Jessika Feliciano, et al., '''ClcR-based biosensing system in the detection of cis-dihydroxylated (chloro-)biphenyls''', ''Analytical and Bioanalytical Chemistry'', 385: 807–813 (2006) | ||
== Results == | == Results == | ||
| + | ===1. Protocols=== | ||
| + | |||
| + | We have made great efforts on refining experiments protocols and shared several protocols that proved to be effecient. Our results show that you will have successful experiments if these protocols were used. | ||
| + | |||
| + | We have shared experience with several other teams, and comunicated a lot. | ||
| + | |||
| + | ===2. Plasmids constructed=== | ||
| + | |||
| + | We have submitted 6 standard parts including dxnA, dbfB, redA2,dxnB, bphB, bphC, bphD. These are genes coding critical enzymens in the degradation pathway of dioxin and PCBs. | ||
| + | |||
| + | ===3. A functional part === | ||
| + | |||
| + | Promoter PpcbC and FYP was fused into a plasmids using pZE12 as the vector. | ||
| + | |||
| + | == Acknowledgments == | ||
| + | |||
| + | *Professor Rolf-Michael wittich and Professor Kenneth N. Timmis, Division of Microbiology, GBF-National Reasearch Centre for Biotechnology, D-38124 Braunschweig, Germany | ||
| + | |||
| + | For the bacteria Sphingomonas sp. Strain RW1 they provided. | ||
| + | |||
| + | * Professor David N. Dowling, Department of Microbiology, University College, Cork, Ireland | ||
| + | |||
| + | For the bacteria E. coil SMl0 provided. | ||
| + | |||
| + | * Professor Junfeng Niu, School of Enviroment, Beijing Normal University | ||
| + | |||
| + | For his instructions and advices | ||
| + | |||
| + | * Dr Yingwu Huang, Bioinformatics Institute, Tsinghua University | ||
| + | |||
| + | For his idea and instructions | ||
| + | |||
| + | * [[Team:Tsinghua|Tsinghua Team]] | ||
| + | |||
| + | For the experience we shared and the indispensable help. | ||
| + | |||
| + | * [[Team:Chiba|Chiba Team]] | ||
| + | |||
| + | For all the information we shared and all the joy we had, especially Mai and Yoshimi. | ||
| + | |||
| + | * [[Team:Tokyo_Tech|Tokyo Tech Team]] | ||
| + | |||
| + | For all the information and advice and encouragement. | ||
| + | |||
| + | * [[Team:USTC|USTC Team]] | ||
| + | |||
| + | For useful suggestions on our experiments and delicious food in Hefei, especially Bo and Jian. | ||
Latest revision as of 04:27, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Notebook |
|---|
Contents |
Overall
We are aiming to create some magic intelligent bacteria to track and ‘eat’ pollutants PCBs (Polychlorinated Biphenyl) and dioxins efficiently, based on the methods of synthetic biology.
Polychlorinated biphenyls (PCBs) are a family of compounds produced commercially by the direct chlorination of biphenyl using ferric chloride and/or iodine as the ctalyst.The total amount of PCBs produced in the world is estimated 1.2 million tons.Because PCBs have been released into the environment in many countries over decades, these compounds have become serious and global environmental contaminants.PCBs tend to accumulate in biota owing to their lypophilic property.
The biphenyl molecule is made up of two connected rings of six carbon atoms each, and a PCB is any molecule having multiple chlorines attached to the biphenyl nucleus.
Two distinct classes of bacteria have now been identified that biodegrade PCBs by different mechanisms, including aerobic bacteria which live in oxygenated environments and anaerobic bacteria which live in oxygen free environments such as aquatic sediments. The aerobes attack PCBs oxidatively , breaking open the carbon ring and destroying the compounds. Anaerobes, on the other hand, leave the biphenyl rings intact while removing the chlorines.
Polychlorinated dibenzo-p-dioxins (PCDD) and polychlorinated dibenzofurans (PCDF) were introduced into the biosphere on a large scale as by-products from the manufacture of chlorinated phenols and the incineration of wastes. Due to their high toxicity they have been the subject of great public and scientific scrutiny.
The evidence in the literature suggests that PCDD/F compounds are subject to biodegradation in the environment as part of the natural chlorine cycle. Lower chlorinated dioxins can be degraded by aerobic bacteria from the genera of Sphingomonas, Pseudomonas and Burkholderia.However, higher chlorinated dioxins requires anaerobic degradation process.
Organic pollutants such as PCB and dioxins, produced in human beings activities in the last century, are toxic and carcinogenic which are able to promulgate widely and accumulate to a high level of concentration by food chain. Due to their inherent thermal and chemical stability, it is commonly considered as indestructible under normal incineration or burial.
Nonetheless, by endowing some bacteria ability of utilizing such molecules as carbon source, cooperative evolution makes all possible! Enzymes assembled from related degradation pathways into our host strain serve as the function part. We introduce popular components involved in chemotaxis, quorum-sensing to regulatory parts, sense the environment signal, respond to move, accelerate growing and produce related degradation enzymes. After the cleaning work being finished, bacteria will return to the normal state.
Taking the condition of our lab into account, we decide just deal with the aerobic degradation path way. And do some work on increasing the degradation effeciency.
Project Details
background information
- Although many environmental pollutants are efficiently degraded by microorganisms, others such as PCBs and dioxins persist and constitute a severe health hazard. In some instances, persistence is a consequence of the inadequate catabolic potential of the available microorganisms. Gene technology, combined with a solid knowledge of catabolic pathways and microbial physiology, enables the experimental evolution of new or improved catabolic activities for such pollutants.
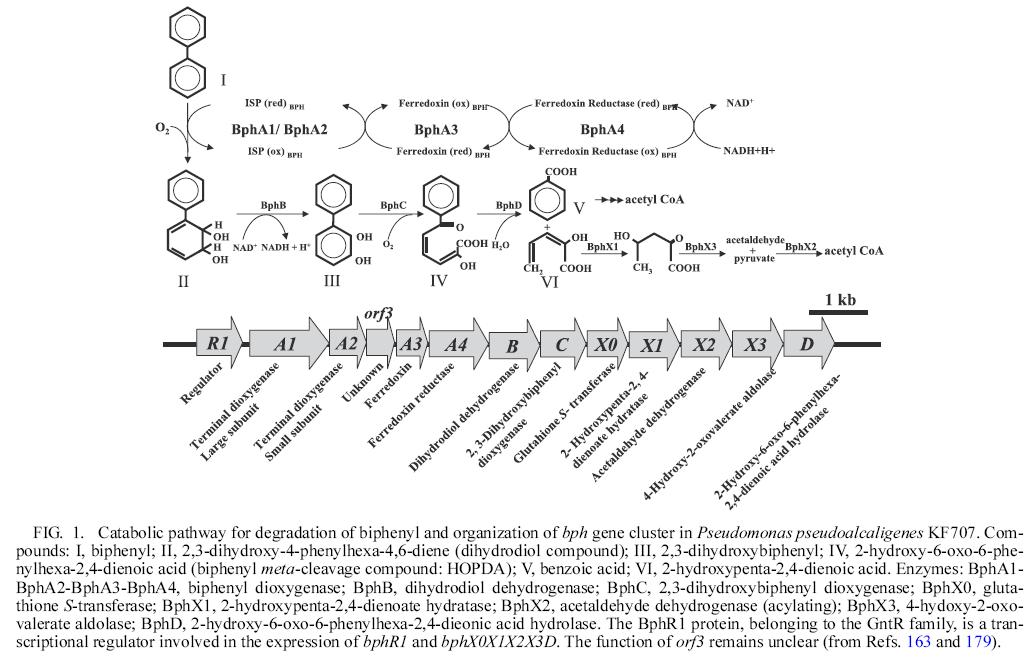
- Catabolic pathway for degradation of biphenyl and organization of bph gene cluster is as follows:
Enzymes responsible for oxidative degradation of PCBs
1 BphA: biphenyl dioxygenase
Biphenyl dioxygenase is a Riesketype three-component enzyme, comprising a terminal dioxygenase that is composed of a large subunit (encoded by bphA1) and a small subunit (encoded by bphA2), a ferredoxin (encoded by bphA3) and a ferredoxin reductase (encodedby bphA4)
It catalyzed the conversion of biphenyl to dihydrodiol compound in step 1.
2 BphB: dihydriol dehydrogenase
In the second step of the upper pathway BphB enzymes catalyzes the conversion of dihidrodiol to dihydroxy compound.
3 BphC: 2,3-dihydroxybiphnyl dioxygenase
The third enzyme in upper pathway is a 2,3-dihyroxybiphnyl dioxygenase involved in the ring meta-cleavage at the 1,2 position.
4 BphD: hydrolase
In the forth step BphD, a hydrolase, hydrolyzes the ring meta-cleavage yellow compound(HOPDA) to chlorobenzoic acid and 2-hydroxypenta-2,4-dienoate.
Design Abstract
Polychlorinated biphenyls (PCBs) are a group of organic pollutants that are persistent when released into the environment. Our task is to design an effective as well as intelligent PCBs degrader. Thus our work could be divided into two parts. One is to develop a sensing system for the detection of PCBs and activation of the downstream degradation pathway; the other is to enhance the degradation efficiency.
DHBD(2,3-dihydroxybiphenyl 1,2-dioxygenase), the third enzyme of the bph pathway, is of particular significance because it is incapable o f catalyzing the ring cleavage step and is subject to various types of inhabitation, as well as suicide inactivation. According to the recent research, ortho-chlorinated PCB metabolites (DHBs) are potent and physiologically significant inhibitors of DHBD, so we design a feedback activation pathway to increase the BphC transcription and expression under a 2, 3-DHBP and 4-CB inducible promoter Ppcb. As 2, 3-DHBP is the second metabolite in the pathway, 2, 3-DHBP at a concentration more than 0.1mM will induce the activate Ppcb and thus increase gene expression downstream.
As dihydrodiols and dihydroxybiphenyls are very toxic to bacterial even after short incubation time, we design a feedback repression pathway use sRNA components— sodB and rhyB. Part of sodB was fusion with bphA and biosurfactant pathway and will be repressed by rhyB which is under the control the promoter Ppcb. As only when rhyB molecules is much more than sodB fusion mRNA, it will play a role of repression, only the high concentration of DHBP will arise this reaction. In this case, the amount of these two metabolites will be reduced in two ways.
As to the sensor part, dihydroxylated PCBs are substrate of the clcA-encoded chlorocatechol dioxygenase and thus induce the clcR and related promoter, so we use this as the sensing system. And T7 amplification system is add downstream to amplify the signal.
The Experiments
- 1.Get the parts from biobrick
We largely follow the instructions provided by the webpage, however, more TE buffer is added(10ul). It seems that this will increase the amount of the plasmids dissolved and improve transformation effeciency.
- 2 PCR
- 2.1 The pfu/Taq complex system
Reagent Concentration/Activity 50ul in total taq buffer 10x 5 pfu/taq complex 0.8~1.0 dNTPmix 10mM each 4 Primer 1 10uM 1.5 Primer 2 10um 1.5 Template DNA changeable -- ddH2O --- add to 50ul
- 2.2 The program under pfu/Tag complex system
Progress Program I
Predenaturing 95℃ 5 min Denaturing 95℃ 30sec Annealing (Tm-5) ℃ 30sec Extension 72℃ theoretically 1min/1kb Last extention 72℃ 5min Hold 4℃
- 3 restriction enzyme digestion
select suitable enzymes and buffer. Analyse the system using NEB cutter in case of double digestion. [http://www.neb.com/nebecomm/DoubleDigestCalculator.asp NEB cutter finder]. 37℃ water bath for 2~3h, 4h is preferable
- 4 ligation
The key to a successful ligation according to our experience is avoiding high temperature. After a gel extraction with 32ul elution buffer, a approximately 20ul product is obtained. Then mixed with 20ul Buffer 1(Takara Ligation Kit). Reaction at 16℃ water bath is widely recommended, and time for ligation we use is 2h or more.
- 5 transformation
Add 10ul ligation product into 100ul competence cells(Top10 or others) and keep it in 4℃ refrigeratory for 30min. After a hotshock of 45sec(for chemical competent) or 90sec(for CaCl2 competent), the cells are placed in the 4℃ refrigeratory again for 3-5min. Then add 600~800ul SOC to hotshock cells and incubate in 37℃ shaking table for 1h(160rpm-180rpm). At last the cells are palced on petri dishes with relative antibiotics at appropriate concentration. Then place in 37℃(temperature accords to the properties of different hosts and plasmids),incubate for 12h or more.
Special protocol
To obtain some genes, we often have to use bacteria chromosome or large size plasmid as template in which high GC% content and complex secondary structures are seriously hampering PCR and thus leading to complete failure. To solve this problem, we have developed a special protocol-- hot start method combined with additive(sole or mixed)-- which is most helpful .
Simplified Hot Start PCR
Hot start method is to prevent primer dimer and low specified product yield. In our experiment, We use common taq polymerase to replace commercial high-priced hot-start polymerase.
After a 5min 95℃ pre-heat step, DNA polymerase is added to each PCR tube before the PCR cycling begin. After that, the regular PCR cycling could begin.
Explanation
When reaction components are mixed at room temperature, reaction set up below the optimal primer annealing temperature, which permits nonspecific primer annealing and extention.Undesired, non-specific primer extetion products formed this way may be amplified in the PCR, resulting in misprimed products and primer ologomers.In hot start PCR, DNA polymerase is withheld from the mixture until the system has reached a temperature that favours specific primer annealing. As a result hot start PCR can greatly improve specificity, sensitivity and yield in a PCR.
Effective Additives
The most simple additive in our PCR is DMSO(>=99.9% purity) at a concentration of 5%(V/V). It works well in most 'problematic' PCR, however, when DMSO fails in some case, we have to turn to a magic mixed additive (2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA) for help. This excellent additive has solved several PCR where even 5% DMSO fails.
In certain PCR there is no yield without this additives, for example the ones by which we amplify bphA1A2A3A4 and bphBC.
Explanation
One major factor limiting the output of PCR routines is that a number of DNA sequences are poorly or not amplifiable under standard reaction conditions, either because of their high GC-content or/and their instrict propertoties to form secondary structures. This protocol is intended for GC-rich DNA sequences. This is a concentration dependent combination of betaine, dithiothreitol, and dimethyl suloxide. According to the references the concentration ratio is: a mixture containing 2.7 M betaine, 6.7 mM DTT, 6.7% DMSO, and 55 ug/ml BSA. It is stable at -20℃ for at least 3 months.
References for special protocol:
1. David E. Birch et al., Simplified hot start PCR, Nature 381:445-446 (1996)
2. S.Kaijalainen et al., An alternative hot start technique for PCR in small volumes using beads of wax-embedded reaction components dried in trehalose, Nucleic Acids Research 21:2959-2960 (1993)
3. Yukihiro Kitade et al., Effect of DMSO on PCR of Porphyra yezoensis(Rhodophyta) gene, Journal of Applied Phycology 15:555-557 (2003)
4. Markus Ralser et al., An efficient and economic enhancer mix for PCR, Biochemical and Biophysical Research Communications 347:747–751 (2006)
Design Details
Enhance the degradation efficiency & detect PCBs
1. Switch on the degradation pathway
The system is awakened by PCBs.
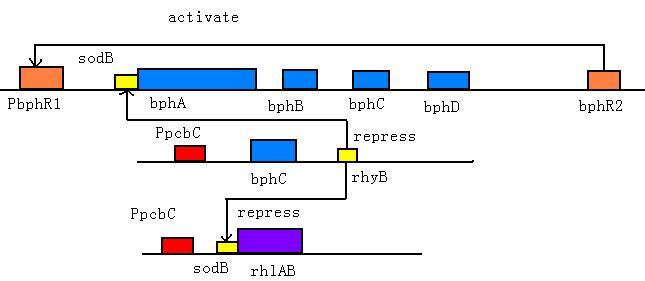
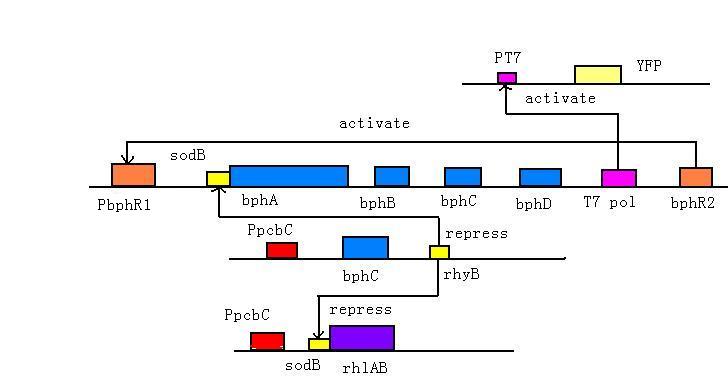
We add bphR1 upstream the pathway and bphR2 downstream. bphR2 could sense the presence of PCBs and bphR1 could sense HOPDA, the third step product. These two regulators could switch on the degradation pathway.
In the absence of PCBs, small amounts of BphR2 protein bind to the bphR2 operator to re press bphR2 transcription (auto-repression). Under this condition, the levels of transcription of bph genes are very low. In the presence of PCBs, the BphR2 protein binds to the operators of bphR1 and bphA1A2A3A4BC and activates their transcription; thus high levels of HOPDA (the ring meta-cleavage compound) are produced from PCBs. In the presence of HOPDA, the transcription of bphR1 is further promoted, and its product BphR1 activates the transcription of bphR1 itself and downstream pathway [1,2].
2. Solve the bottleneck in PCBs degradation pathway
Our idea: Add a spur track to solve the bottleneck of the pump.
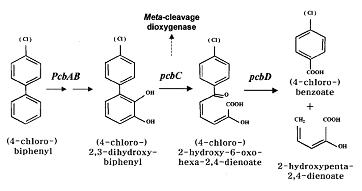
BphC (2,3-dihydroxybiphenyl 1,2-dioxygenase), the third enzyme of the bph pathway, is of particular significance because it is incapable o f catalyzing the ring cleavage step and is subject to various types of inhabitation, as well as suicide inactivation. According to the recent research, ortho-chlorinated PCB metabolites (DHBs) are potent and physiologically significant inhibitors of BphC, so we design a feedback activation pathway using small RNA to increase the BphC transcription and expression under a 2, 3-DHBP and 4-CB inducible promoter Ppcb [3]. As 2, 3-DHBP is the second metabolite in the pathway, 2, 3-DHBP at a concentration more than 0.1mM will induce the activate PpcbC and thus increase gene expression downstream. The activated threshold of PpcbC is relatively high, so this pathway will be activated when main PCBs degradation above has began.
3. Control on the amount of PCBs molecular that enter the cell.
As dihydrodiols (first step product) and dihydroxybiphenyls (DHBP, second step product) are very toxic to bacterial even after short incubation time, we design a feedback repression pathway use sRNA components— sodB and rhyB. Part of sodB was fusion with bphA and biosurfactant pathway and will be repressed by rhyB[4] which is under the control the promoter Ppcb. As only when rhyB molecules is much more than sodB fusion mRNA, it will play a role of repression, only the high concentration of DHBP will arise this reaction. This design will control the amount of PCBs molecular that enter through the membrane.
4. System guarantee
In our design T7 amplification system is used to amplify the signal. T7 polymerase is added downstream the main degradation pathway. When T7 polymerase is translated, it will activate the T7 promoter and downstream reporter gene, YFP. Then the signal could be detected. This system could be used to test the function of the degradation pathway.
References for Design Details:
[1] Kensuke Furukawa, et al., Microbial Degradation of Polychlorinated Biphenyls: Biochemical and Molecular Features, Journal of Bioscience and Bioengineering 105:433–449 (2008)
[2] Stephen Y. K. Seah, et al., Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls, Journal of Bacteriology 183:1511–1516 (2001)
[3] Sang-ho Park, et al., Construction of transformant reporters carrying fused genes using pcbC promoter of Pseudomonas sp DJ-12 for detection of aromatic pollutants, Environmental Monitoring and Assessment 92:241–251 (2004)
[4] Qinhong Wang, et al., Engineering Bacteria for Production of Rhamnolipid as an Agent for Enhanced Oil Recovery, Biotechnology and Bioengineering, 98:842-853 (2007)
[5] Jessika Feliciano, et al., ClcR-based biosensing system in the detection of cis-dihydroxylated (chloro-)biphenyls, Analytical and Bioanalytical Chemistry, 385: 807–813 (2006)
Results
1. Protocols
We have made great efforts on refining experiments protocols and shared several protocols that proved to be effecient. Our results show that you will have successful experiments if these protocols were used.
We have shared experience with several other teams, and comunicated a lot.
2. Plasmids constructed
We have submitted 6 standard parts including dxnA, dbfB, redA2,dxnB, bphB, bphC, bphD. These are genes coding critical enzymens in the degradation pathway of dioxin and PCBs.
3. A functional part
Promoter PpcbC and FYP was fused into a plasmids using pZE12 as the vector.
Acknowledgments
- Professor Rolf-Michael wittich and Professor Kenneth N. Timmis, Division of Microbiology, GBF-National Reasearch Centre for Biotechnology, D-38124 Braunschweig, Germany
For the bacteria Sphingomonas sp. Strain RW1 they provided.
- Professor David N. Dowling, Department of Microbiology, University College, Cork, Ireland
For the bacteria E. coil SMl0 provided.
- Professor Junfeng Niu, School of Enviroment, Beijing Normal University
For his instructions and advices
- Dr Yingwu Huang, Bioinformatics Institute, Tsinghua University
For his idea and instructions
For the experience we shared and the indispensable help.
For all the information we shared and all the joy we had, especially Mai and Yoshimi.
For all the information and advice and encouragement.
For useful suggestions on our experiments and delicious food in Hefei, especially Bo and Jian.
 "
"