Team:Cambridge/Bacillus
From 2008.igem.org
| Line 52: | Line 52: | ||
* Pxyl - xylose inducible, present on ''B. subtilis'' vector ECE153 | * Pxyl - xylose inducible, present on ''B. subtilis'' vector ECE153 | ||
| - | Information about the ''B. subtilis'' vectors can be found [ | + | Information about the ''B. subtilis'' vectors can be found [[Team:Cambridge/Signalling/Vectors here]. |
===The Schematic:=== | ===The Schematic:=== | ||
Revision as of 03:35, 30 October 2008
|
x
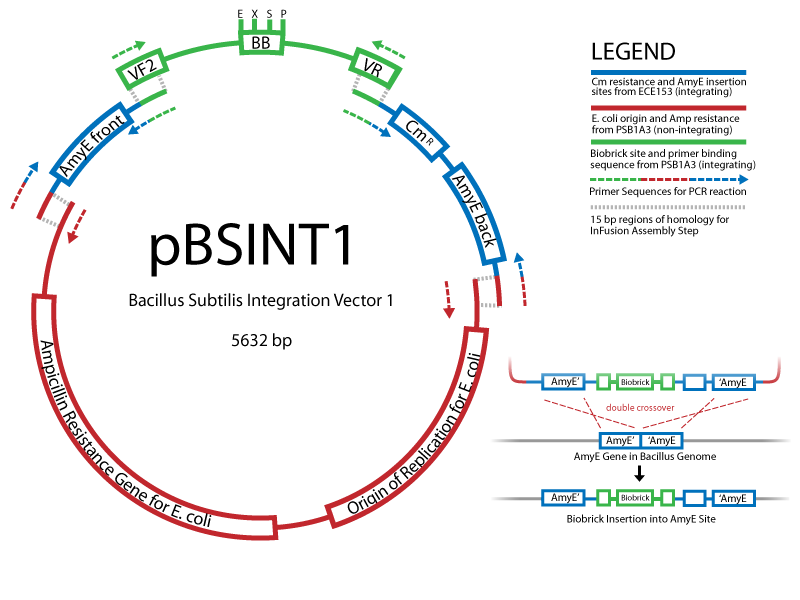
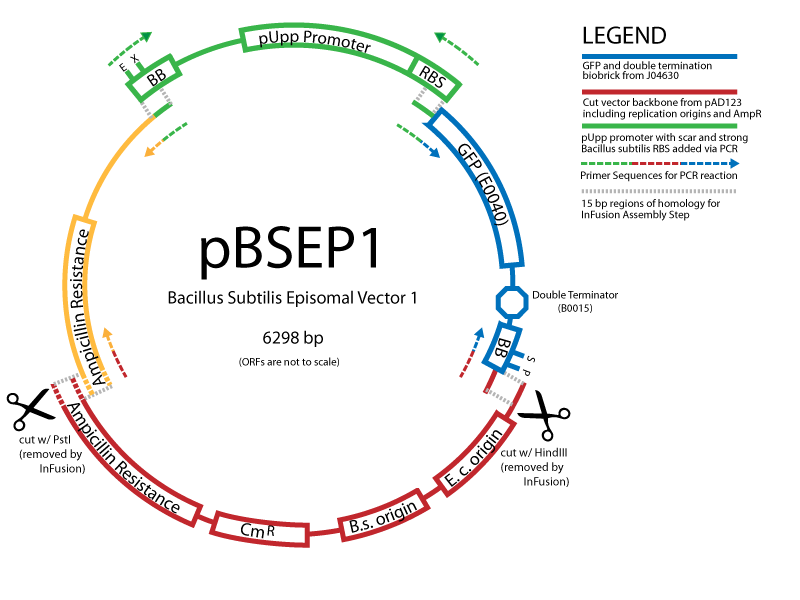
Working with B.subtilisBuilding on the work of the last year's Cambridge iGEM team, we are exploring applications of the gram-positive chassis B.subtillis. Easy to handle and transform, this bacterium is much better to use than E.coli wherever protein import/export is concerned, e.g. in our signalling project. An important part of our effort is to establish standard protocols and parts to work in B.subtillis, characterise control elements, and develop new vectors. You can find our Bacillus protocols here. Creating new Biobrick-compatible B.subtilis vectors[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Vectors submitted:] We are using the In-Fusion™ PCR method from ClonTech to create new Biobrick-compatible integrative and episomal vectors. These vectors will allow us to insert Biobricks into the Bacillus subtilis genome at the AmyE locus and as a 3-5 copy plasmid that will replicate autonomously in Bacillus.
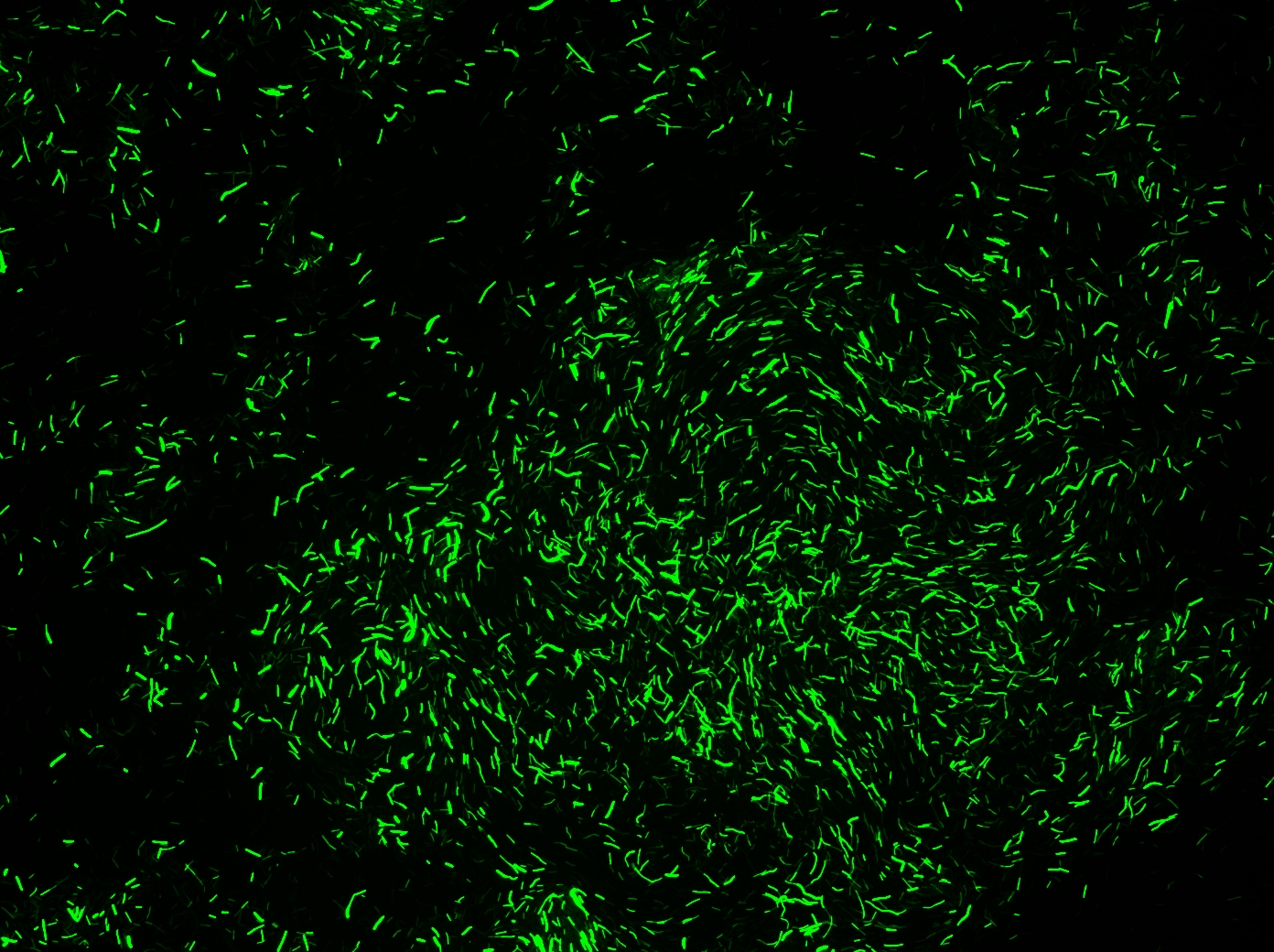
Successful transformation of BacillusTransformation with episomal vectorsECE166 : High-level constitutive expression of a Green Fluorescent Protein (with the promoter Pupp) Transformation with integration vectorsECE153 and ECE112 : Transformation in IA751 : Amylase test B. subtilis Promoters Designed: [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge Biobricks submitted:]
Information about the B. subtilis vectors can be found [[Team:Cambridge/Signalling/Vectors here]. The Schematic:
x
|
 "
"