Team:ETH Zurich/Wetlab/Genome Reduction
From 2008.igem.org
(→Lab results) |
|||
| Line 24: | Line 24: | ||
| - | [[Image:proof_of_concept_construct,_symbols.jpg|center|thumb|500px|1 | + | [[Image:proof_of_concept_construct,_symbols.jpg|center|thumb|500px|Figure 1: Construct for proof of concept]] <br clear="all" /> |
| Line 34: | Line 34: | ||
| - | [[Image:proof_of_concept,_symbols.jpg|center|thumb|500px|2 | + | [[Image:proof_of_concept,_symbols.jpg|center|thumb|500px|Figure 2: Proof of concept]] <br clear="all" /> |
| Line 55: | Line 55: | ||
| - | [[Image:modified_proof_of_concept_construct,_symbols.jpg|center|thumb|300px|3 | + | [[Image:modified_proof_of_concept_construct,_symbols.jpg|center|thumb|300px|Figure 3: Modified construct for proof of concept]]<br clear="all" /> |
| Line 63: | Line 63: | ||
| - | [[Image:modified_proof_of_concept,_symbols.jpg|center|thumb|300px|4 | + | [[Image:modified_proof_of_concept,_symbols.jpg|center|thumb|300px|Figure 4: Modified proof of concept]]<br clear="all" /> |
==== SceI restriction enzyme ==== | ==== SceI restriction enzyme ==== | ||
| Line 77: | Line 77: | ||
For the proof of concept we want to clone the following constructs: | For the proof of concept we want to clone the following constructs: | ||
| - | [[Image:SceI_RFP_and_T4_SceI,_final.png|center|thumb|500px|5 | + | [[Image:SceI_RFP_and_T4_SceI,_final.png|center|thumb|500px|Figure 5: Plasmids for proof of concept]]<br clear="all" /> |
These two plasmids are going to be transduced into the same host. | These two plasmids are going to be transduced into the same host. | ||
Revision as of 03:07, 30 October 2008
|
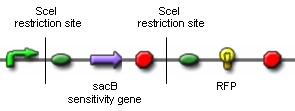
Genome reductionGoalIn this section we discuss about how to randomly delete chromosomal fragments of E. coli in order to reduce the physical amount of genomic DNA. MethodTo reach our goal of random deletion of chromosomal fragments, we want to synthesize a frequently cutting restriction enzyme along with the simultaneous, shortly delayed, or even continuous synthesis of a ligase. The restriction enzyme will cut genomic DNA in a random fashion in vivo, while the ligase performs its job of religation. Assuming that the genomic DNA is cut at several sites within one cell, religation will lead to the exclusion of chromosomal fragments in some cells. Multiple rounds of restriction and religation will therefore lead to a markedly reduced genome. Proof of conceptIn 1997 Ren et al. showed that in vivo religation of linearized vector and insert is possible by overexpression of the T4 ligase (1). Additionally, Heitman et al. proposed that intracellular DNA which has been cut in vivo by EcoRI is religated by the ligase T4 (2). Our idea, however, relies on the assumption that in vivo restriction and religation is possible and leads to the exclusion of chromosomal DNA. We are trying to verify these assumptions in several experiments which will from now on be refered to as our “proof of concept”. Our proof of concept relies on the following construct (not shown are ribosomal binding sites behind both SceI restriction sites and a terminator following the RFP):
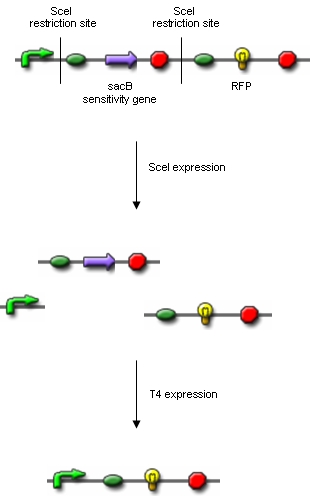
Additionally, we ordered DNA encoding the T4 ligase and the SceI restriction enzyme. Transformation of these plasmid-encoded enzymes into cells carrying the proof of concept construct is supposed to yield bacteria that can be induced to express T4 and SceI. Before synthesis of T4 and SceI, cells carrying the proof of concept construct do not synthesize RFP. After synthesis of the restriction enzyme, SceI will cut the bacterial chromosome at the sites indicated above. The ligase will then religate the construct, leading to the exclusion of the sensitivity gene and the terminator in some of the cells. Hence, these cells would synthesize the RFP reporter protein and could easily be identified:
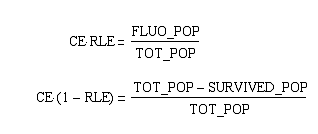
Extracting modelling parameters from the proof of concept experimentA very important parameter in our reduction strategy approach is undoubtly the efficiency on which the process of cutting, fragment removal and re-ligation is going to occure in-vivo. Our proof of concept construct and experiment were designed in order to provide us two important indexes of efficiency, namely the cutting efficiency (CE) and the removal and ligation efficiency (RLE). The CE parameter represents the probability that at least one cut is made into the chromosome of a cell for a pulse of expression of the restriction enzyme. The RLE represents the probability that after a cutting a fragment is lost and the chromosome is relagated. This efficiency frequencies are going to help us understand the behaviour of the single cell when expression of the restriction enzyme is induced. In order to calculate the two parameters from the experiment we planned to:
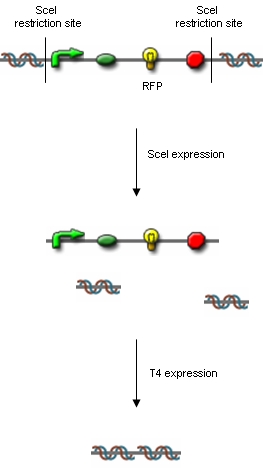
From these three population counts we can set up the system of two equations: After solving the equations we can use the parameters for modelling the probability of one fragment to be removed, as we used in the modelling section Genome Scale Model. Modification of proof of conceptUnfortunately, up to now we have not received our construct for the proof of concept. Therefore, we are working on a modified construct:
After transformation of bacteria with the construct shown above and plasmids containing the genes coding for SceI restriction enzyme and the T4 ligase, induction of gene expression will lead to in vivo cutting and religation of the RFP-encoding plasmid, leading to the loss of the RFP gene from the plasmid. Therefore, if cells lose the RFP signal after induction of the restriction enzyme and ligase, we have proven that in vivo restriction and ligation is possible and can lead to the exclusion of DNA fragments:
SceI restriction enzymeSceI is a site-specific homing endonuclease. It is extremely rare-cutting as it recognizes an 18 bp sequence. These properties make SceI a perfect restriction enzyme for our proof of concept for which the restriction enzyme should only cut the sites of our constructs indicated above. However, for our goal of minimizing E. coli’s genome we will need to use a frequently cutting restriction enzyme which will potentially lead to severe damage of the cell’s genome. In order to limit this damage, we want to be able to pulse the expression of the restriction enzyme. Therefore, we are cloning an inducible promoter in front of SceI. T4 ligaseT4 is the commonly used ligase for in vitro cloning. For in vivo cloning, high levels of T4 are advantageous not only for improving the efficiency of religation leading to the exclusion of chromosomal fragments, but also for limiting DNA damage. However, constitutive overexpression of T4 might lead to immediate religation without the exclusion of chromosomal fragments. Therefore, we are cloning T4 behind an IPTG-inducible promoter (R0010), and, as an alternative approach, behind several constitutive promoter of differing strengths. Another idea would be to clone both SceI and T4 behind the same promoter, so that induction would lead to simultaneous expression of both enzymes. Lab resultsGoals For the proof of concept we want to clone the following constructs: These two plasmids are going to be transduced into the same host. The RFP generator enclosed from two SceI-restriction sites will be on a low-copy plasmid, like pCK01, to reduce the number of cutting targets for SceI. Whereas T4 ligase and SceI restriction enzyme will be placed on a high-copy plasmid. Expression of T4 and SceI will be induced in order to cut out the whole RFP generator and religate the digested plasmid in vivo. This will be easily detected by the disappearance of the red fluorescence of the cells. We have chosen to use RFP, since they are easily distinguishable on plates from the ones which dont express RFP anymore. By using antibiotics it will be easily assured that only colonies which contain the plasmids with the two resistance genes will survive. Modified proof of conceptFor the RFP generator we used the Biobrick BBa_J04450 and cloned SceI-restriction sites into its EcoRI- and PstI-site. picture showing BBaJ04450, indicating of E and P-site where oligos with SceI-site are inserted. For the SceI-restriction sites we ordered oligos containing the SceI-recognition site and at its ends the overhangs for EcoRI- and PstI-site respectively. The oligos were hybridised and cloned into BBa_J04450. Successful cloning were verified by digestion and gel electrophoresis. Oligos for SceI-site integrated at EcoRI-site: EcoR_SceI_up
AATTCAGTTACGCTAGGGATAACAGGGTAATATAGC
EcoR_SceI_down
AATTGCTATATTACCCTGTTATCCCTAGCGTAACTG
Pst_site_up
AGTTACGCTAGGGATAACAGGGTAATATAGCTGCA
Pst_site_down
GCTATATTACCCTGTTATCCCTAGCGTAACTTGCA
picture with hybridised oligos showing overhangs, killed site, and SceI-recognition site SceI restriction enzyme and T4 ligaseWe performed following clonings to bring SceI and T4 under the control of different promoters by standard Biobrick assembly:
References(1) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T39-3V4BTWM-CH&_user=791130&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_version=1&_urlVersion=0&_userid=791130&md5=5f7b6177feaf9c0d06cd0e9b334a43ce Ren Z. J., Baumann R. G., Black L. W. (1997): Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: construction of a T4 phage hoc gene display vector. Gene 22 195 (2):303-11.] (2) [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=2648397 Heitman J., Zinder N. D., Model P. (1989): Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci 86 (7):2281-5.] (3) [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=10829079 Datsenko K. A., Wanner B. L. (2000): One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97 (12):6640-5.] |
 "
"