Team:Illinois/Antibody Receptor Tyrosine Kinase Fusion Notebook
From 2008.igem.org
(Difference between revisions)
(→October 2, 2008) |
(→October 24, 2008) |
||
| Line 748: | Line 748: | ||
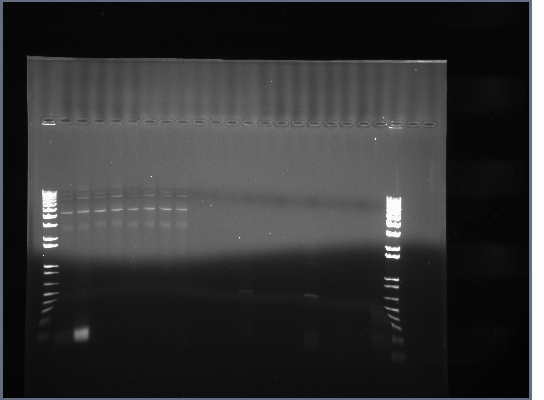
* Control transformation using XL1 Blue cells from mutagenesis kit worked perfectly w/ pUC18, pSB1A7, and pSB1A3. These cells were kept in the -80degree freezer the whole time and were commercially prepared, further implying that our competent cells aren't competent at all. | * Control transformation using XL1 Blue cells from mutagenesis kit worked perfectly w/ pUC18, pSB1A7, and pSB1A3. These cells were kept in the -80degree freezer the whole time and were commercially prepared, further implying that our competent cells aren't competent at all. | ||
| + | |||
| + | [[Image:PA270209.JPG|600px]] | ||
* Made new batch of competent cells, see 7-18-08 for procedure. | * Made new batch of competent cells, see 7-18-08 for procedure. | ||
Revision as of 02:34, 30 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
July 1, 2008
Made Yeast Media
- YPD Medium, per liter:
- 10g yeast extract
- 20g peptone
- 20g dextrose
- 20g agar(only for plates)
- Dextrose filter sterilized, the rest autoclaved
- Weigh nutrients into flask double the volume you want to make, and stir to dissolve
- Dextrose added to autoclaved media to equivalent of 20 g/L
- Liquid media placed on bench, plate media placed in 65 degree water bath approximately 5 minutes
- Poured into plates and allowed to solidify
July 17, 2008
E.Coli Media
- LB medium, per liter
- 10g tryptone
- 5g yeast extract
- 5g NaCl
- 1 mL 1N NaOH
- 15g (agar for plates)
- Procedure:
- Antibody DNA resuspended in TE buffer, 0.1 μg/μL
- 5mL LB inoculated with single colony DH5a pro
- Incubated at 37 degrees overnight
July 18, 2008
- We made competent cells to use for transformations.
- Procedure:
- 3mL overnight culture of DH5a pro
- Inoculated into 35mL of LB
- OD600 checked, want 0.2-0.3
- Place culture on ice for 3 minutes
- Spin at 10,000 rpm for 7 minutes, discard supernatant
- Resuspend in 10 mL cold 30 mM CaCl2, spin as before
- Resuspend in 3 ml cold 30 mM Cacl2, incubated on ice overnight
- Glycerol added to 10%, cells in freezer
July 19, 2008
- Transformation: We received the synthesized TE33 antibody genes from IDT on plasmids. We transformed these plasmids into DH5a cells and plated on LB+amp to screen for transformants.
- Cells put on ice 30 min with 1μL plasmid(100ng)
- Heat Shock for 2 minutes at 42 degrees
- Ice for 8 minutes
- Add cells to 1ml LB
- Grow for 1 hour
- Plate 100 to 200 μL on LB amp, grow overnight
July 21, 2008
- Observations:
- There were plenty of transformants on the plates. 5ml of LB+amp was inoculated for plasmid extraction the next day.
July 22, 2008
- Mini spin prep kit was used to extract the plasmids carrying the TE33 light and heavy chains. From these plasmids we can run PCRs to amplify the regions of the antibody chains.
- DNA eluted in 100μL H2O and frozen.
July 29, 2008
- All primers brought to standard concentration, 30mM
- 33.3μL H2O added per mole of primer
July 30, 2008
- Assembly PCR: The antibody genes synthesized by IDT contained the entire antigen binding fragments of the light and heavy chain, but to construct a single chain antibody we only need the variable regions of these genes. This PCR will amplify just those sections of the gene and will leave both the light and heavy chain fragments with complementary ends that can be assembled in a following PCR.
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Antibody chain | H | H | L | L | H | H | L | L |
| DNA source | mini-prep | synthesized IDT genes | ||||||
| template | 0.5ul | |||||||
| primers | 1ul of appropriate forward and reverse primer | |||||||
| MgCl2 | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul |
| master mix | 20ul | |||||||
| H20 | to 50ul total volume | |||||||
- Master mix already contains MgCl2; should have added extra to some tubes.
- PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
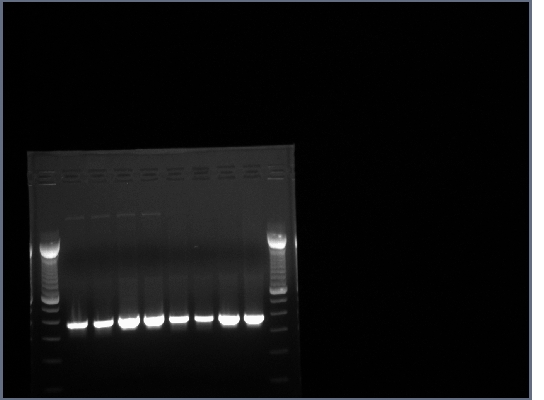
- 1.5% agarose gel made, 0.75g agarose in 50ml 0.5X TBE w/10ul EtBr.
- 20ul of PCR produts loaded on gel w/4ul 6X loading buffer, run at 200V for about half an hour.
- All lanes had lots of DNA at ~375bp which is the expected length, PCR worked.
July 31, 2008
- Another PCR to asemble the TE33 heavy and light chain fragments from yesterday. The complementary region codes for a flexible glycine-serine linker that will join the two halves of the single chain antibody.
| tube | 1 | 2 | 3 | 4 | ||||
| template from 7-30 PCR | 2+3 | 2+7 | 6+7 | 1+8 | ||||
| template | 0.5ul of both heavy and light chains | |||||||
| primers | 1ul of appropriate forward and reverse primer | |||||||
| MgCl2 | 0ul | 0ul | 3ul | 3ul | ||||
| master mix | 20ul | |||||||
| H20 | to 50ul total volume | |||||||
- PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- 72 degrees 5 min
- HOLD at 4 degrees
- Products run on a 1.5% agarose gel, no products in the expected 700-800bp range.
August 1, 2008
- PCR troubleshooting of yesterday's reaction:
- Use gradient for annealing temp (40-65 degrees), use more template, leave out end primers
- Four series of reactions using a gradient annealing temp were run in addition to a fifth series omitting the end primers.
| reaction series | A | B | C | D | E |
| template (5+7) from 7-30 PCR | 0.5ul | 1ul | 1.5ul | 2ul | 1ul |
| primers | 1ul of forward and reverse primers (omitted in E series) | ||||
| master mix | 20ul | ||||
| H20 | to 50ul total volume | ||||
- Annealing temps: 40, 43.8, 50, 54, 60, and 64 degrees
- PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- annealing step 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
- PCR products run on 1.5% agarose gel at 200V for ~20 minutes, then 240V for ~20 minutes.
- Products in the 700-800bp range in all lanes. Perhaps it worked because of using a differnt product from the 7-30 PCR. We now have the DNA for our single chain antibody.
August 6, 2008
- 1.5% agarose gel of 8-1 PCR products run to cut out bands. Products A1, B1, C1, and E1 used. We will use this purified DNA for making biobricks and future PCRs to assemble our antibody-receptor fusion.
- Bands around 800bp cut out for all lanes and invitrogen gel extraction kit used to collect DNA in 50ul H2O.
September 5, 2008
- Flk-1 clones from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones ATCC] streaked onto LB+amp and incubated at 37 degrees.
- Flk-1 is a mouse gene coding for a vascular endothelial growth factor receptor, a tyrosine kinase receptor. We chose this particular receptor because it is well characterized and because it appears to activate simply through dimerization. This is important because if ligand binding caused a conformational change that helped to activate the receptor it is unlikely that dimerization caused by the single-chain antibody binding cholera toxin would create a similar conformational change.
September 6, 2008
- Colonies picked from flk-1 plate and inoculated into 5ml LB+amp, grown at 37 degrees.
- Freeze-dried e. coli with YCp50-poly from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=87555&Template=vectors ATCC] resuspended in 5ml LB, grown at 37 degrees.
- YCp50-poly is the yeast/E. coli shuttle vector we chose to place our receptor gene on. It allows for selection using ampicilin in E. coli and media lacking uracil in yeast.
September 7, 2008
- Mini spin prep of flk-1 and YCp50-poly cells, plasmids stored in 50ul H2O.
- We now have purified supplies of the plasmid carrying the flk-1 gene and our shuttle vector.
September 25, 2008
- Making biobricks
- Here we ran PCRs to include the biobrick prefix and suffix sequences into the genes for the TE33 heavy and light chains, the single-chain antibody, and flk-1. PCRs were also run to amplify various lengths of the flk-1 gene. We want to investigate whether certain sections of the extracellular domain of flk-1 contribute to dimerization activity, or if much of the extracellular domain can be replaced by the single-chain antibody. Flk-1 links 1, 2, and 3 delete 222, 420, and 660 amino acids from the N terminus of flk-1 respectively. These deletions were chosen to conserve varying numbers of immunoglobulin domains found in the extracellular portion of flk-1. Structural information can be found [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR009136 here].
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 0.5uL template | H chain | L chain | SCA | SCA | SCA | SCA | flk-1 | flk-1 |
| IDT | IDT | A | B | C | E | A | B | |
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
| tube | 9 | 10 | 11 | 12 | 13 | 14 | ||
| 0.5uL template | SCA w/ link | SCA w/ link | SCA w/ link | SCA w/ link | flk link 1 | flk link 1 | ||
| A | B | C | E | A | B | |||
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
| tube | 15 | 16 | 17 | 18 | ||||
| 0.5uL template | flk link 2 | flk link 2 | flk link 3 | flk link 3 | ||||
| A | B | A | B | |||||
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
- PCR program
- 94 degrees 4 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 3 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
September 26, 2008
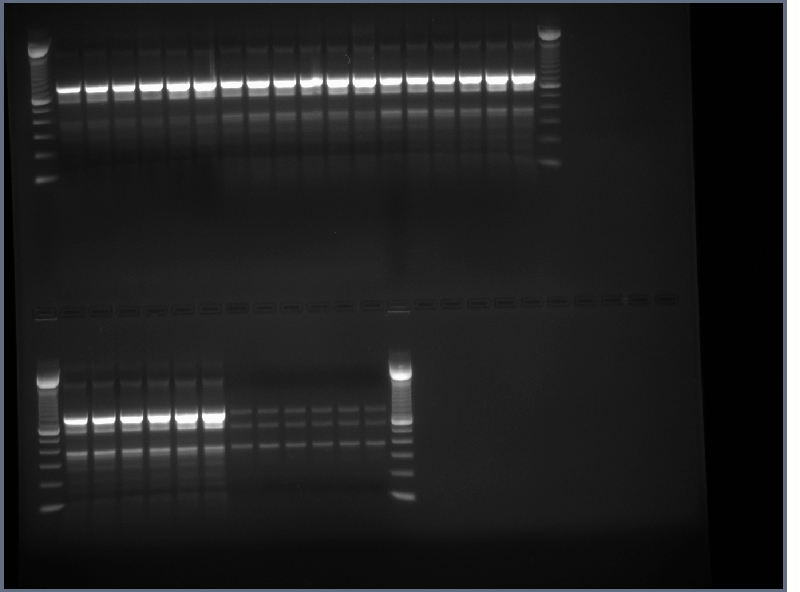
- Product from yesterday run on 1% gel, 200V for ~30 minutes
- The results were mostly as expected. The biobricks for the light and heavy antibody chains, and the single chain antibody were of the expected lengths. The amount of DNA for flk-1 based of products seems to be low, and it is hard to tell if the bands are of the proper size because they are larger than the lader that was used.
- Lanes 1,2,6,8,12,14,16,18 were cut out and purified with invitrogen gel extraction kit, eluted in 50ul H2O.
- Although it is unclear whether the flk-1 PCRs worked, we will continue on and run a PCR to assemble the the single-chain antibody with the flk-1 fragments. We will be using a similar strategy as we used to assemble the single-chain antibody: the ends of each fragment will have complementary sequences which will anneal and amplify in the PCR reaction to yield a full length product. The linker region will again code for a flexible glycine-serine linker, but the sequence was rearranged to avoid any incorrect annealing with the linker region already present in the single-chain antibody.
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| template | 0.5ul flk-1 A | 0.5ul flk-1 B | 0.5ul flk link 1 A | 0.5ul flk link 1 B | 0.5ul flk link 2 A | 0.5ul flk link 2 B | 0.5ul flk link 3 A | 0.5ul flk link 3 B | 0.5ul SCA link + 0.5ul flk link 1 | 0.5ul SCA link + 0.5ul flk link 1 (crude) | 0.5ul SCA link + 2ul flk link 1 | 0.5ul SCA link + 2ul flk link 1 (crude) | 0.5ul SCA link + 0.5ul flk link 2 | 0.5ul SCA link + 0.5ul flk link 2 (crude) | 0.5ul SCA link + 2ul flk link 2 | 0.5ul SCA link + 2ul flk link 2 (crude | 0.5ul SCA link + 0.5ul flk link 3 | 0.5ul SCA link + 0.5ul flk link 3 (crude) | 0.5ul SCA link + 2ul flk link 3 | 0.5ul SCA link + 2ul flk link 3 (crude) |
| primers | 1uL forward & reverse for all | |||||||||||||||||||
| mastermix | 8.25uL for all | |||||||||||||||||||
| H2O | to 50ul total volume | |||||||||||||||||||
- PCR program
- 94 degrees 4 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 3 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
September 27, 2008
- Running another PCR reaction to attempt to get better amplification of flk-1.
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| template | 0.5ul flk-1 A | 0.5ul flk-1 B | 0.5ul flk link 1 A | 0.5ul flk link 1 B | 0.5ul flk link 2 A | 0.5ul flk link 2 B | 0.5ul flk link 3 A | 0.5ul flk link 3 B |
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | to 50ul total volume | |||||||
- PCR program
- 94 degrees 4 min
- 94 degrees 1 min
- 58 degrees 1 min
- 72 degrees 3 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
- 0.8% agarose gel run of products from yesterday, 200V for ~45 minutes
- The first 8 reactions may have worked. The band around 4500bp is the proper length for the full length flk-1 gene, but again it is not very bright. Also, the different length flk-1 fragmetns all seem to be running at approximately the same size on the gel.
September 28, 2008
- 0.8% gel run of yesterday's products, 150 V, 38 min, 200 V, 10 min
- The bands are still very dim and the fragments are not running at the correct length.
September 30, 2008
- Now we will try to transform a biobrick plasmid to insert our parts into so that we can send them to the registry.
- p1010 plasmid for biobricks cut out of plate 1003, well 4A; soaked in 5uL warm TE (50°C) for ~20 min, put on ice with 100ul DH5a for 30 min, heat shocked at 50°C for 60s, placed on ice 2 min, 200uL LB added, incubated at 37°C 1 hour, plated on LB + Amp and incubated overnight.
- It turns out the above transformation was a stupid thing to do as it contained the cell death gene. Another transformation with part 1008:1C was carried out.
| tube | 1 | 2 | 3 | 4 | 5 | 6 |
| template | A=0.5uL B=1.0uL C=1.5uL | |||||
| primers | 1uL forward & reverse for all | |||||
| mastermix | 8.25uL for all | |||||
| H2O | A=39.25uL B=38.75ul C=38.25ul | |||||
- gradient annealing temperatures of 55.0, 57.8, 59.3, 61.0, 63.5, 65.0 were used
- Run for 39 cycles
October 2, 2008
- PCR products from yesterday run on 0.8% gel, 200V, ~40min
- Similar results as before, the PCR of flk-1 and flk-1 fragments is proving rather troublesome.
- pSB1A3 & pSB1A7 cut out of registry binder and transformed using the supplied protocol.
- Plated on LB+Amp
October 3, 2008
- PCR from yesterday may have kind of worked, just not a lot of product, running another gel to find out
- 10-2 Transformation failed, transforming w/plasmid containing flk-1 to test transformation protocol
- Hard to tell if PCR products are correct length, so-so bands
October 4, 2008
- transformation with the flk-1 plasmid worked well, there were plenty of colonies
- our transformation procedure probably works fine and the competency of our cells is ok
October 8, 2008
- Perhaps the spots in the registry binder we have been using are bad, will try transforming other parts.
- Transformations of parts 1003:2F, 1008:10D, 1008:1C, 1008:6G
October 9, 2008
- A few colonies were seen on the plates of the transformations from 10-8 but not a lot. We will do a plasmid miniprep followed by another transformation to see if these colonies really have the plasmid.
October 10, 2008
- plasmid miniprep using 5 Prime Spin Mini kit, plasmid eluted in 50ul H2O and stored in the freezer
October 14, 2008
- Setup transformations using plasmid from 10/10/2008 to check whether or not we really had transformants
October 16, 2008
- Transformation on 10-10 failed
- next we'll try using Top10 chemically competent cells from invitrogen rather then the DH5a cells we made ourselves
- 50uL cells transformed with 1003:10D, 1003:2F, plasmid from 10/10/2008, and pUC19 control
- another PCR to atempt to make the antibody-receptor fusion
| tube | 1 | 2 | 3 | 4 | 5 |
| template | 0.5uL SCA + 0.5uL flk-link3 | 0.5uL SCA + 1.0uL flk-link3 | 0.5uL SCA + 1.5uL flk-link3 | 0.5uL SCA + 2.0uL flk-link3 | 0.5uL SCA + 1.5uL flk-link3 |
| primers | 1.0uL forward & reverse | none | |||
| mastermix | 20.0uL for all | ||||
| H2O | 27.0uL | 26.5uL | 26.0uL | 25.5uL | 28.0uL |
October 17, 2008
- 10-16 transformation failed. The Top10 cells from invitrogen may have lost competency as they were incorrectly stored in a -20 degree freezer instead of the -80 degree and could have been thawed and refrozen.
- 1% gel of yesterday's PCR run, 200V, ~40min, no bands
October 18, 2008
- Because our antibody-receptor fusion gene is fairly long (4kb) We will now try to use a high fidelity polymerase (phusion).
| tube | 1 | 2 | 3 | 4 | 5 |
| template | 0.5uL SCA + 0.5uL flk-link3(8) | 0.5uL SCA + 1.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) | 0.5uL SCA + 2.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) |
| primers | 1.0uL forward & reverse | no primers | |||
| buffer | 10.0uL for all | ||||
| phu | 0.5uL for all | ||||
| H2O | 36.5uL | 36.0uL | 35.5uL | 35.0uL | 37.5uL |
- bands present in 3 of the 5 lanes but still not of the right length
October 21, 2008
- Due to a lack of time to continually troubleshoot transformations, we requested strains carrying pSB1A7 and pSB1A3 from igem HQ (thanks!). We used these frozen stocks LB+Amp cultures for plasmid prep.
October 22, 2008
- Performed plasmid miniprep on cultures from yesterday
- Next we setup restriction digests of the freshly isolated plasmids and our biobricks so we can ligate them together and transform into bacteria.
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 5uL DNA | pSB1A7 | pSB1A3 | H chain brick | L chain brick | SCA brick | flk-1 brick | flk-1 plasmid |
| 5uL buffer2 | |||||||
| 0.5uL BSA | |||||||
| 1uL RE 1 | EcoRI | ||||||
| 1uL RE 2 | PstI | HindIII | |||||
| 37.5uL H20 | |||||||
October 23, 2008
- Innoculated 5mL culture with DHSα PRO to make a fresh batch of competent cells tomorrow.
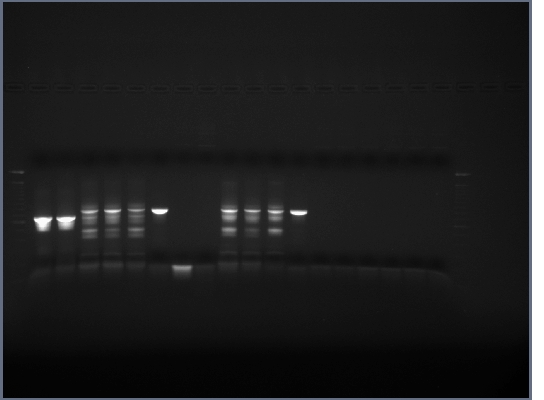
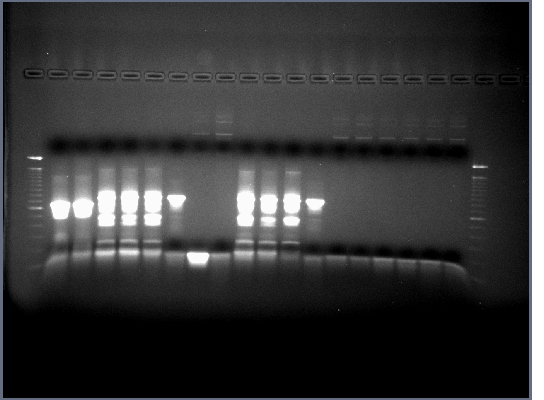
- Ran 0.8% gel of restriction digests, no band in H brick lane (this gene contains an internal EcoRI restriction site that we didn't have time to mutate out) or flk-1 brick (expected as no flk-1 PCR ever worked too well), the rest of the bands were as expected.
- Cut out and purified bands for ligations and PCR.
- PCR to get try to get flk-1 fragments using gel purified restriction product
- 0.5mL purified flk-1 fragment, 1uL F & R primers, 20uL mastermix, 27.5uL H20
- primers to amplify full lenght flk-1 as well as flk-1 link 1, 2, and 3
October 24, 2008
- Control transformation using XL1 Blue cells from mutagenesis kit worked perfectly w/ pUC18, pSB1A7, and pSB1A3. These cells were kept in the -80degree freezer the whole time and were commercially prepared, further implying that our competent cells aren't competent at all.
- Made new batch of competent cells, see 7-18-08 for procedure.
- Ligation of restriction products from yesterday:
- 2uL vector, 6uL insert, 0.5mL T4 ligase, 1uL buffer, room temp 1 hr
- Flk-1 PCR from yesterday failed.
- DHSα PRO transformed w/ pSB1A7 + L chain, pSB1A7 + SCA, pSB1A3 + L chain, pSB1A3 + SCA
October 25, 2008
- Transformations failed, forgot to run a control so could be because the newly made competent cells are bad. Ligation may have been unsuccessful also.
- Transformed XL1 Blue w/ 5uL of ligation products pSB1A7 + L chain, pSB1A7 + SCA, pSB1A3 + L chain, pSB1A3 + SCA, and also 1uL pUC18.
- Repeated ligation using more DNA and trying rapid ligation protocol:
- 3uL vector, 9uL insert, 4uL buffer, 1uL T4 ligase, 3uL H20, room temp 5 min
- Used 1uL of new ligation to transform XL1 Blue and 2uL to transform DHSα PRO
October 26, 2008
- Transformation failed (except controls)
- Setting up more transformations, pSB1A7 + L, pSB1A7 + SCA, pSB1A7, pUC18 in Top10 and DHSα
October 27, 2008
- restriction digest of L chain and SCA using more DNA to hopefully improve the ligation, same procedure as 10-22-08 except 10ul of DNA
- trying electroporation instead of chemical transformation
- 1ul of 10-25 ligation in 50ul of electrocompetent DH5a
- put 50ul in electroporation cuvette being careful not to get bubbles
- tap cuvette so that cells are even
- insert into electroporater and press pulse button
- recover 1 hour in 1ml of SOC and then plate
- Troubleshooting tips from Joel
- cells probably suck
- try variable heat shock (30 sec, 45 sec, etc)
- run digest >6 hours, check on gel and purify
- run ligation >6 hours at 20 degrees or RT, can refrigerate and use ligation again the next day, clean up ligation reaction, run ligase/no ligase gels, 4:1 insert:vector molar ratio (16:1 volume according to Rob)
October 28, 2008
- one colony on plate from pSB1A7 transformation
- picked colony and put in 100ul DMSO for colony PCR, also started 5ml overnight culture
- colony PCR run, didn't appear to be a band in the 600bp range, maybe just an empty vector transformation
October 29, 2008
- plasmid miniprep run on overnight culture
- possible L chain biobrick sent to igem
 "
"